Sulfonamide derivatives as Mycobacterium tuberculosis inhibitors: in silico approach
- PMID: 30607317
- PMCID: PMC6314710
- DOI: 10.1007/s40203-018-0041-9
Sulfonamide derivatives as Mycobacterium tuberculosis inhibitors: in silico approach
Abstract
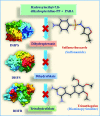
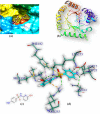
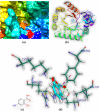
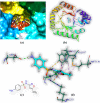
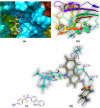
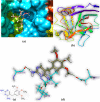
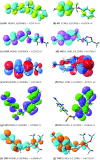
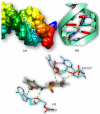
Both DHPS (dihydropteroate synthase) and DHFR (dihydrofolate reductase) play important physiological roles in the survivability of Mycobacterium tuberculosis (MTB). Sulfonamides are the potent drugs to monitor growth and proliferation of MTBs by inhibiting the activity of DHPS and DHFR which could explain the mechanism of action of these molecules. In this work, 102 heterocyclic sulfonamides (HSF) have been screened by discovery studio molecular docking programme to search the best suitable molecule for the treatment of MTBs. Lipinski's rule of five protocols is followed to screen drug likeness of these molecules and ADMET (absorption, distribution, metabolism, excretion and toxicity) filtration has been used to value their toxicity. Only fourteen molecules are found to obey the Lipinski's rule and able to cross the ADMET filter. A small difference between HOMO and LUMO energy signifies the electronic excitation energy which is essential to calculate molecular reactivity and stability of the best docked compound and easy activation of drug in the protein environment. Both 4-amino-N-(6-hydroxypyridin-2-yl)benzenesulfonamide (M1) and 4-amino-N-(9H-carbazol-2-yl)benzenesulfonamide (M2) show the best theoretical efficiency with DHPS and DHFR, respectively. These compounds are also found to bind to the adenine-thymine region of tuberculosis DNA.
Keywords: ADMET; DHPS and DHFR inhibitors; Heterocyclic sulfonamide compounds; Molecular docking; Structure based drug design.
Conflict of interest statement
Compliance with ethical standardsThe authors confirm that this article content has no conflicts of interest.
Figures

References
-
- Becke AD. Density-functional thermochemistry. I. The role of exact exchange. J Chem Phys. 1993;98(7):5648–5652. doi: 10.1063/1.464913. - DOI
LinkOut - more resources
Full Text Sources

