Quantitative Proteomic Analysis of Small and Large Extracellular Vesicles (EVs) Reveals Enrichment of Adhesion Proteins in Small EVs
- PMID: 30608700
- PMCID: PMC6411421
- DOI: 10.1021/acs.jproteome.8b00647
Quantitative Proteomic Analysis of Small and Large Extracellular Vesicles (EVs) Reveals Enrichment of Adhesion Proteins in Small EVs
Abstract
Extracellular vesicles (EVs) are important mediators of cell-cell communication due to their cargo content of proteins, lipids, and RNAs. We previously reported that small EVs (SEVs) called exosomes promote directed and random cell motility, invasion, and serum-independent growth. In contrast, larger EVs (LEVs) were not active in those assays, but might have unique functional properties. In order to identify protein cargos that may contribute to different functions of SEVs and LEVs, we used isobaric tags for relative and absolute quantitation (iTRAQ)-liquid chromatography (LC) tandem mass spectrometry (MS) on EVs isolated from a colon cancer cell line. Bioinformatics analyses revealed that SEVs are enriched in proteins associated with cell-cell junctions, cell-matrix adhesion, exosome biogenesis machinery, and various signaling pathways. In contrast, LEVs are enriched in proteins associated with ribosome and RNA biogenesis, processing, and metabolism. Western blot analysis of EVs purified from two different cancer cell types confirmed the enrichment of cell-matrix and cell-cell adhesion proteins in SEVs. Consistent with those data, we found that cells exhibit enhanced adhesion to surfaces coated with SEVs compared to an equal protein concentration of LEVs. These data suggest that a major function of SEVs is to promote cellular adhesion.
Keywords: adhesion; exosomes; extracellular vesicles; iTRAQ; microvesicles; proteomics.
Conflict of interest statement
Conflict of interest statement
No potential conflicts of interest were disclosed.
Figures

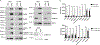

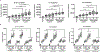
References
-
- van Niel G; D’Angelo G; Raposo G, Shedding light on the cell biology of extracellular vesicles. Nature Reviews Molecular Cell Biology 2018, 19, (4), 213–228. - PubMed
-
- Zhang H; Freitas D; Kim HS; Fabijanic K; Li Z; Chen H; Mark MT; Molina H; Martin AB; Bojmar L; Fang J; Rampersaud S; Hoshino A; Matei I; Kenific CM; Nakajima M; Mutvei AP; Sansone P; Buehring W; Wang H; Jimenez JP; Cohen-Gould L; Paknejad N; Brendel M; Manova-Todorova K; Magalhaes A; Ferreira JA; Osorio H; Silva AM; Massey A; Cubillos-Ruiz JR; Galletti G; Giannakakou P; Cuervo AM; Blenis J; Schwartz R; Brady MS; Peinado H; Bromberg J; Matsui H; Reis CA; Lyden D, Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field-flow fractionation. Nat Cell Biol 2018, 20, (3), 332–343. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

