Differential and convergent utilization of autophagy components by positive-strand RNA viruses
- PMID: 30608919
- PMCID: PMC6334974
- DOI: 10.1371/journal.pbio.2006926
Differential and convergent utilization of autophagy components by positive-strand RNA viruses
Abstract
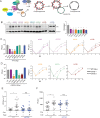
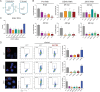
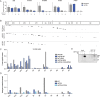
Many viruses interface with the autophagy pathway, a highly conserved process for recycling cellular components. For three viral infections in which autophagy constituents are proviral (poliovirus, dengue, and Zika), we developed a panel of knockouts (KOs) of autophagy-related genes to test which components of the canonical pathway are utilized. We discovered that each virus uses a distinct set of initiation components; however, all three viruses utilize autophagy-related gene 9 (ATG9), a lipid scavenging protein, and LC3 (light-chain 3), which is involved in membrane curvature. These results show that viruses use noncanonical routes for membrane sculpting and LC3 recruitment. By measuring viral RNA abundance, we also found that poliovirus utilizes these autophagy components for intracellular growth, while dengue and Zika virus only use autophagy components for post-RNA replication processes. Comparing how RNA viruses manipulate the autophagy pathway reveals new noncanonical autophagy routes, explains the exacerbation of disease by starvation, and uncovers common targets for antiviral drugs.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

