Transcriptomic characterization of signaling pathways associated with osteoblastic differentiation of MC-3T3E1 cells
- PMID: 30608923
- PMCID: PMC6319725
- DOI: 10.1371/journal.pone.0204197
Transcriptomic characterization of signaling pathways associated with osteoblastic differentiation of MC-3T3E1 cells
Abstract
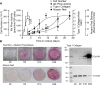
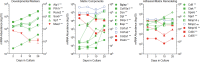
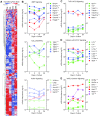
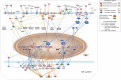
Bone remodeling involves the coordinated actions of osteoclasts, which resorb the calcified bony matrix, and osteoblasts, which refill erosion pits created by osteoclasts to restore skeletal integrity and adapt to changes in mechanical load. Osteoblasts are derived from pluripotent mesenchymal stem cell precursors, which undergo differentiation under the influence of a host of local and environmental cues. To characterize the autocrine/paracrine signaling networks associated with osteoblast maturation and function, we performed gene network analysis using complementary "agnostic" DNA microarray and "targeted" NanoString nCounter datasets derived from murine MC3T3-E1 cells induced to undergo synchronized osteoblastic differentiation in vitro. Pairwise datasets representing changes in gene expression associated with growth arrest (day 2 to 5 in culture), differentiation (day 5 to 10 in culture), and osteoblast maturation (day 10 to 28 in culture) were analyzed using Ingenuity Systems Pathways Analysis to generate predictions about signaling pathway activity based on the temporal sequence of changes in target gene expression. Our data indicate that some pathways involved in osteoblast differentiation, e.g. Wnt/β-catenin signaling, are most active early in the process, while others, e.g. TGFβ/BMP, cytokine/JAK-STAT and TNFα/RANKL signaling, increase in activity as differentiation progresses. Collectively, these pathways contribute to the sequential expression of genes involved in the synthesis and mineralization of extracellular matrix. These results provide insight into the temporal coordination and complex interplay between signaling networks controlling gene expression during osteoblast differentiation. A more complete understanding of these processes may aid the discovery of novel methods to promote osteoblast development for the treatment of conditions characterized by low bone mineral density.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






Similar articles
-
BMP-2 controls alkaline phosphatase expression and osteoblast mineralization by a Wnt autocrine loop.J Bone Miner Res. 2003 Oct;18(10):1842-53. doi: 10.1359/jbmr.2003.18.10.1842. J Bone Miner Res. 2003. PMID: 14584895
-
Galectin-3 is essential for proper bone cell differentiation and activity, bone remodeling and biomechanical competence in mice.Metabolism. 2018 Jun;83:149-158. doi: 10.1016/j.metabol.2018.02.001. Epub 2018 Feb 9. Metabolism. 2018. PMID: 29432728
-
Distinct proliferative and differentiated stages of murine MC3T3-E1 cells in culture: an in vitro model of osteoblast development.J Bone Miner Res. 1992 Jun;7(6):683-92. doi: 10.1002/jbmr.5650070613. J Bone Miner Res. 1992. PMID: 1414487
-
Ets transcription factors and targets in osteogenesis.Oncogene. 2000 Dec 18;19(55):6455-63. doi: 10.1038/sj.onc.1204037. Oncogene. 2000. PMID: 11175361 Review.
-
Genetic and transcriptional control of bone formation.Oral Maxillofac Surg Clin North Am. 2010 Aug;22(3):283-93, v. doi: 10.1016/j.coms.2010.05.001. Oral Maxillofac Surg Clin North Am. 2010. PMID: 20713262 Free PMC article. Review.
Cited by
-
G Protein-Coupled Receptor Systems and Their Role in Cellular Senescence.Comput Struct Biotechnol J. 2019 Aug 23;17:1265-1277. doi: 10.1016/j.csbj.2019.08.005. eCollection 2019. Comput Struct Biotechnol J. 2019. PMID: 31921393 Free PMC article. Review.
-
Physiological Mineralization during In Vitro Osteogenesis in a Biomimetic Spheroid Culture Model.Cells. 2022 Aug 30;11(17):2702. doi: 10.3390/cells11172702. Cells. 2022. PMID: 36078105 Free PMC article.
-
DPP4-Truncated CXCL12 Alters CXCR4/ACKR3 Signaling, Osteogenic Cell Differentiation, Migration, and Senescence.ACS Pharmacol Transl Sci. 2022 Dec 13;6(1):22-39. doi: 10.1021/acsptsci.2c00040. eCollection 2023 Jan 13. ACS Pharmacol Transl Sci. 2022. PMID: 36659961 Free PMC article.
-
The Highly Potent AhR Agonist Picoberin Modulates Hh-Dependent Osteoblast Differentiation.J Med Chem. 2022 Dec 22;65(24):16268-16289. doi: 10.1021/acs.jmedchem.2c00956. Epub 2022 Dec 2. J Med Chem. 2022. PMID: 36459434 Free PMC article.
-
Non-canonical Wnt signaling participates in Jagged1-induced osteo/odontogenic differentiation in human dental pulp stem cells.Sci Rep. 2022 May 9;12(1):7583. doi: 10.1038/s41598-022-11596-9. Sci Rep. 2022. PMID: 35534526 Free PMC article.
References
-
- Raisz LG. Physiology and pathophysiology of bone remodeling. Clin Chem. 1999;45:1353–8. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

