The Ribosome as a Missing Link in Prebiotic Evolution III: Over-Representation of tRNA- and rRNA-Like Sequences and Plieofunctionality of Ribosome-Related Molecules Argues for the Evolution of Primitive Genomes from Ribosomal RNA Modules
- PMID: 30609737
- PMCID: PMC6337102
- DOI: 10.3390/ijms20010140
The Ribosome as a Missing Link in Prebiotic Evolution III: Over-Representation of tRNA- and rRNA-Like Sequences and Plieofunctionality of Ribosome-Related Molecules Argues for the Evolution of Primitive Genomes from Ribosomal RNA Modules
Abstract
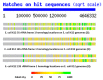
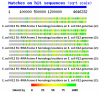
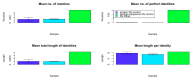
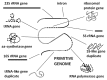
We propose that ribosomal RNA (rRNA) formed the basis of the first cellular genomes, and provide evidence from a review of relevant literature and proteonomic tests. We have proposed previously that the ribosome may represent the vestige of the first self-replicating entity in which rRNAs also functioned as genes that were transcribed into functional messenger RNAs (mRNAs) encoding ribosomal proteins. rRNAs also encoded polymerases to replicate itself and a full complement of the transfer RNAs (tRNAs) required to translate its genes. We explore here a further prediction of our "ribosome-first" theory: the ribosomal genome provided the basis for the first cellular genomes. Modern genomes should therefore contain an unexpectedly large percentage of tRNA- and rRNA-like modules derived from both sense and antisense reading frames, and these should encode non-ribosomal proteins, as well as ribosomal ones with key cell functions. Ribosomal proteins should also have been co-opted by cellular evolution to play extra-ribosomal functions. We review existing literature supporting these predictions. We provide additional, new data demonstrating that rRNA-like sequences occur at significantly higher frequencies than predicted on the basis of mRNA duplications or randomized RNA sequences. These data support our "ribosome-first" theory of cellular evolution.
Keywords: LINE; SINE; aminoacyl tRNA synthetase; extra-ribosomal; genome; mRNA; non-canonical; paralog; rRNA; ribosome; ribosome-binding protein; tRNA.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
The ribosome as a missing link in prebiotic evolution II: Ribosomes encode ribosomal proteins that bind to common regions of their own mRNAs and rRNAs.J Theor Biol. 2016 May 21;397:115-27. doi: 10.1016/j.jtbi.2016.02.030. Epub 2016 Mar 4. J Theor Biol. 2016. PMID: 26953650
-
The ribosome as a missing link in the evolution of life.J Theor Biol. 2015 Feb 21;367:130-158. doi: 10.1016/j.jtbi.2014.11.025. Epub 2014 Dec 10. J Theor Biol. 2015. PMID: 25500179
-
Accretion history of large ribosomal subunits deduced from theoretical minimal RNA rings is congruent with histories derived from phylogenetic and structural methods.Gene. 2020 May 15;738:144436. doi: 10.1016/j.gene.2020.144436. Epub 2020 Feb 3. Gene. 2020. PMID: 32027954
-
What do we know about ribosomal RNA methylation in Escherichia coli?Biochimie. 2015 Oct;117:110-8. doi: 10.1016/j.biochi.2014.11.019. Epub 2014 Dec 13. Biochimie. 2015. PMID: 25511423 Review.
-
Biosynthesis and function of posttranscriptional modifications of transfer RNAs.Annu Rev Genet. 2012;46:69-95. doi: 10.1146/annurev-genet-110711-155641. Epub 2012 Aug 16. Annu Rev Genet. 2012. PMID: 22905870 Review.
Cited by
-
Circular code motifs in the ribosome: a missing link in the evolution of translation?RNA. 2019 Dec;25(12):1714-1730. doi: 10.1261/rna.072074.119. Epub 2019 Sep 10. RNA. 2019. PMID: 31506380 Free PMC article.
-
Rapid and accurate identification of ribosomal RNA sequences via deep learning.Nucleic Acids Res. 2022 Jun 10;50(10):e60. doi: 10.1093/nar/gkac112. Nucleic Acids Res. 2022. PMID: 35188571 Free PMC article.
-
Interchangeable utilization of metals: New perspectives on the impacts of metal ions employed in ancient and extant biomolecules.J Biol Chem. 2021 Dec;297(6):101374. doi: 10.1016/j.jbc.2021.101374. Epub 2021 Oct 31. J Biol Chem. 2021. PMID: 34732319 Free PMC article. Review.
-
RNP-world: The ultimate essence of life is a ribonucleoprotein process.Genet Mol Biol. 2022 Sep 23;45(3 Suppl 1):e20220127. doi: 10.1590/1678-4685-GMB-2022-0127. eCollection 2022. Genet Mol Biol. 2022. PMID: 36190700 Free PMC article.
-
Emergence of a "Cyclosome" in a Primitive Network Capable of Building "Infinite" Proteins.Life (Basel). 2019 Jun 18;9(2):51. doi: 10.3390/life9020051. Life (Basel). 2019. PMID: 31216720 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

