SNF1-Related Protein Kinases SnRK2.4 and SnRK2.10 Modulate ROS Homeostasis in Plant Response to Salt Stress
- PMID: 30609769
- PMCID: PMC6337402
- DOI: 10.3390/ijms20010143
SNF1-Related Protein Kinases SnRK2.4 and SnRK2.10 Modulate ROS Homeostasis in Plant Response to Salt Stress
Abstract
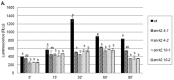
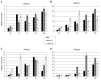
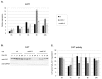
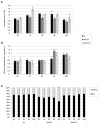
In response to salinity and various other environmental stresses, plants accumulate reactive oxygen species (ROS). The ROS produced at very early stages of the stress response act as signaling molecules activating defense mechanisms, whereas those produced at later stages in an uncontrolled way are detrimental to plant cells by damaging lipids, DNA, and proteins. Multiple systems are involved in ROS generation and also in ROS scavenging. Their level and activity are tightly controlled to ensure ROS homeostasis and protect the plant against the negative effects of the environment. The signaling pathways responsible for maintaining ROS homeostasis in abiotic stress conditions remain largely unknown. Here, we show that in Arabidopsis thaliana, two abscisic acid- (ABA)-non-activated SNF1-releted protein kinases 2 (SnRK2) kinases, SnRK2.4 and SnRK2.10, are involved in the regulation of ROS homeostasis in response to salinity. They regulate the expression of several genes responsible for ROS generation at early stages of the stress response as well as those responsible for their removal. Moreover, the SnRK2.4 regulate catalase levels and its activity and the level of ascorbate in seedlings exposed to salt stress.
Keywords: Arabidopsis thaliana; SnRK2; antioxidant enzymes; ascorbate cycle; hydrogen peroxide; reactive oxygen species; salinity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

