A Novel Partitivirus in the Hypovirulent Isolate QT5-19 of the Plant Pathogenic Fungus Botrytis cinerea
- PMID: 30609795
- PMCID: PMC6356794
- DOI: 10.3390/v11010024
A Novel Partitivirus in the Hypovirulent Isolate QT5-19 of the Plant Pathogenic Fungus Botrytis cinerea
Abstract
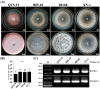
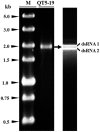
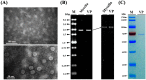
A pink isolate (QT5-19) of Botrytis cinerea was compared with three gray isolates of B. cinerea for growth and morphogenesis on potato dextrose agar (PDA), and for pathogenicity on tobacco. A double-stranded (ds) RNA mycovirus infecting QT5-19 was identified based on its genome feature and morphology of the virus particles. The results showed that QT5-19 grew rapidly and established flourishing colonies as the gray isolates did. However, it is different from the gray isolates, as it failed to produce conidia and sclerotia asthe gray isolates did. QT5-19 hardly infected tobacco, whereas the gray isolates aggressively infected tobacco. Two dsRNAs were detected in QT5-19, dsRNA 1 and dsRNA 2, were deduced to encode two polypepetides with homology to viral RNA-dependent RNA polymerase (RdRp) and coat protein (CP), respectively. Phylogenetic analysis of the amino acid sequences of RdRp and CP indicated that the two dsRNAs represent the genome of a novel partitivirus in the genus Alphapartitivirus, designated here as Botrytis cinerea partitivirus 2 (BcPV2). BcPV2 in QT5-19 was successfully transmitted to the three gray isolates through hyphal contact. The resulting BcPV2-infected derivatives showed rapid growth on PDA with defects in conidiogenesis and sclerogenesis, and hypovirulence on tobacco. This study suggests that BcPV2 is closely associated with hypovirulence of B. cinerea.
Keywords: Botrytis cinerea; conidiogenesis; hypovirulence; partitivirus; sclerogenesis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

