A cis-acting bidirectional transcription switch controls sexual dimorphism in the liverwort
- PMID: 30609993
- PMCID: PMC6418429
- DOI: 10.15252/embj.2018100240
A cis-acting bidirectional transcription switch controls sexual dimorphism in the liverwort
Abstract
Plant life cycles alternate between haploid gametophytes and diploid sporophytes. While regulatory factors determining male and female sexual morphologies have been identified for sporophytic reproductive organs, such as stamens and pistils of angiosperms, those regulating sex-specific traits in the haploid gametophytes that produce male and female gametes and hence are central to plant sexual reproduction are poorly understood. Here, we identified a MYB-type transcription factor, MpFGMYB, as a key regulator of female sexual differentiation in the haploid-dominant dioicous liverwort, Marchantia polymorpha MpFGMYB is specifically expressed in females and its loss resulted in female-to-male sex conversion. Strikingly, MpFGMYB expression is suppressed in males by a cis-acting antisense gene SUF at the same locus, and loss-of-function suf mutations resulted in male-to-female sex conversion. Thus, the bidirectional transcription module at the MpFGMYB/SUF locus acts as a toggle between female and male sexual differentiation in M. polymorpha gametophytes. Arabidopsis thaliana MpFGMYB orthologs are known to be expressed in embryo sacs and promote their development. Thus, phylogenetically related MYB transcription factors regulate female gametophyte development across land plants.
Keywords: Marchantia polymorpha; R2R3 MYB‐type transcription factor; antisense transcription; lncRNA; sexual differentiation.
© 2018 The Authors. Published under the terms of the CC BY 4.0 license.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

- A, B
Development of X chromosome‐containing female (A) and Y chromosome‐containing male (B) M. polymorpha plants.
- C
Development of embryo sac and pollen, in bisexual flowers of A. thaliana.
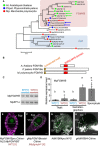
Phylogenetic tree of R2R3‐MYB proteins of clades 11, 12, and 14–16, as described by Bowman et al (2017), from representative land plant species constructed using the maximum‐likelihood method based on conserved MYB domain sequences. See Source Data for the sequences used and accession numbers. Numbers at nodes indicate bootstrap values calculated from 1,000 replicates. The tree is drawn to scale, with branch lengths reflecting the number of substitutions per site. Scale bar, 0.5 substitutions per site. Arrows indicate FGMYB orthologs involved in embryo sac development in Arabidopsis thaliana, MpFGMYB of Marchantia polymorpha (this study), and the most similar Physcomitrella patens genes shown in (B). The FGMYB clade is shaded in pink, and a distantly related FOUR LIPS (FLP) clade is in blue.
Schematic representations of the FGMYB polypeptide structures. R2R3 MYB domains are shown in red and a conserved amino‐terminal motif of PpFGMYBs and MpFGMYB in orange.
Genomic PCR analysis indicating the existence of MpFGMYB in both male [Y] and female [X] genomes of M. polymorpha. Two biological replicates were analyzed. Autosomal MpEF1α was used as a control.
Real‐time RT–PCR analyses indicating preferential accumulation of MpFGMYB transcripts in female sexual organs and the sporophytes. MpEF1α was used for normalization. Measurements of six biological replicates for thalli and sporophyte, and three biological replicates for gametangiophore are plotted. Bars represent mean ± SD. Symbols above the bars indicate grouping by P < 0.05 in a Tukey–Kramer test.
A transcriptional reporter with 5′‐ and 3′‐flanking sequences revealed transcription of MpFGMYB throughout mature archegonia. Scale bar, 10 μm. Magenta, chlorophyll autofluorescence; green, Citrine fluorescence.
MpFGMYB‐Citrine fusion proteins expressed using the 5′‐ and 3′‐flanking sequences rescued the Mpfgmyb ge ‐1 mutant and accumulated in the nuclei of the egg and the ventral canal cell (VCC; Shimamura, 2016). Scale bar, 10 μm. Magenta, chlorophyll autofluorescence; green, Citrine fluorescence.
A transcriptional AtMYB64 reporter (AtMYB64‐NLS‐YFP‐GUS (NYG)) is specifically expressed in all four cell types of the A. thaliana embryo sac (Rabiger & Drews, 2013; Waki et al, 2013). Scale bar, 25 μm. Green, YFP fluorescence; white, cell walls.
Expression of AtMYB64‐Citrine fusion proteins under the AtMYB64 promoter was detected in the central cells (CC) and egg cells (EC) of the A. thaliana embryo sac (Rabiger & Drews, 2013). Scale bar, 25 μm. Green, Citrine fluorescence; white, cell walls.
- A
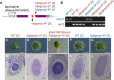
MpFGMYB gene structure and locations of Mpfgmyb mutations. Gray line, 5′‐ and 3′‐flanking sequences; light purple box, UTR; dark purple box, coding region; red box, MYB domain‐coding region; arrowheads, mutation positions; black arrow, transcriptional direction; dotted line, splice patterns.
- B
Diagnosis of genetic sex using Y chromosome‐linked and X chromosome‐linked rbm27 and rhf73 markers, respectively. Two biological replicates were analyzed for each genotype.
- C–L
Gametangiophore morphology (C–G) and gamete development (H–L) of wild‐type and mutant plants. Scale bars, 5 mm (C–G), 10 μm (H and J), 100 μm (I, K, and L).

- A
A schematic representation of the process of archegoniophore morphogenesis after far‐red irradiation. Longitudinal sections of apical notch regions are presented. Around 14 days after induction, dome‐shaped archegoniophore primordia developed at the ventral side of apical notch area. Regions corresponding to the images shown in (B–D) are boxed.
- B–D
Confocal microscopic images of the apical notch region of gMpFGMYBresist‐Citrine plants without (B) or with far‐red irradiation (C, D). MpFGMYB‐Citrine does not accumulate in the apical notch region of vegetative thalli (B). After 10 days of FR irradiation, MpFGMYB‐Citrine accumulates in the ventral side of the apical notch region (C). Expression domain of MpFGMYB‐Citrine expands when the morphology of archegoniophore primordia becomes evident (D). Yellow dotted lines delineate the edges of thalli and a developing archegoniophore. Top, bright field images; middle, fluorescent images; bottom, merged images. Scale bar, 25 μm.
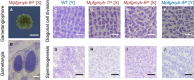
- A
Gross morphology of antheridiophores developed in Mpfgmyb‐6 ge [X].
- B–J
Histological analyses indicating antheridium formation (B), diagonal cell division of spermatogenous cells (C–F), and subsequent spermiogenesis (G–J) in the wild‐type (C, G), two independent Mpfgmyb [X] mutants (D, E, H, and I), and one Mpfgmyb [Y] mutant (F, J).

- A–C
DAPI‐staining visualization of sperm formation in wild‐type male (A), Mpfgmyb [X] (B), and Mpfgmyb [Y] (C) plants. Note that background DAPI staining visualizes flagella (arrows) in addition to nuclei (arrowheads). (B′) is an enlarged image of the boxed region in (B), visualizing an incompletely condensed nucleus. Scale bar, 5 μm.
- D
RT–PCR analysis indicating acquisition of male‐like autosomal gene expression patterns in Mpfgmyb [X] antheridiophores. Two independent Mpfgmyb [X] mutant alleles were analyzed.
- E, F
TEM analyses visualizing the abnormal arrangement of axonemal microtubules in Mpfgmyb [X] sperm (F), as compared with those of wild‐type males (E). Scale bar, 100 nm.

- A
RT–PCR analysis demonstrating loss of expression of female‐specific autosomal genes in the antheridiophores of Mpfgmyb [X]. Note that X chromosome‐linked genes are still expressed in Mpfgmyb [X] antheridiophores as in the wild‐type females, despite their male‐like sexual morphologies. Biological duplicates were analyzed for each genotype.
- B–D
Hoechst‐stained wild‐type archegonia treated with sperm from wild‐type (B), Mpfgmyb [X] (C), and Mpfgmyb [Y] (D) plants, indicating the inability of Mpfgmyb [X] sperm to enter wild‐type archegonia. Arrowheads, sperm in archegonial cavity. Dotted lines, egg cells. Scale bars, 10 μm (B′–D′, B″–D″), 50 μm (B–D).

- A
RNA‐seq analysis showing male‐specific accumulation of lncRNAs derived from the MpFGMYB 3′ region (top), and diagrams illustrating wild‐type and mutant MpFGMYB/SUF loci (bottom). Folded lines with a ∆ symbol indicate a deletion.
- B
RT–PCR analysis of wild‐type and genetically male suf mutants revealed loss of SUF expression and gain of MpFGMYB expression in suf mutants after induction of reproductive growth by far‐red irradiation. Two independent suf mutant alleles were analyzed. The SUF primer pair used here flanked an intron and the duplicated bands of SUF likely represent spliced and unspliced forms.
- C–H
Gametangiophore morphology (C, E and G) and gametangium development (D, F and H) of plants with the designated genotypes. Scale bars, 1 mm (C, E and G), 20 μm (D), 50 μm (F), 100 μm (H).
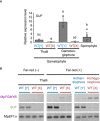
Real‐time RT–PCR analyses indicating preferential accumulation of SUF transcripts in male reproductive organs. Constitutively expressed MpEF1α was used for normalization. Measurements of six biological replicates for thalli and sporophyte, and three biological replicates for gametangiophores are plotted. Bars represent mean ± SD. Symbols above bars indicate grouping by P < 0.05 in a Tukey–Kramer test. See Source Data online for measurements and statistics.
Strand‐specific RT–PCR confirmed male‐specific accumulation of SUF transcripts in vegetative and reproductive organs, regardless of the induction of reproductive growth by far‐red irradiation. Biological duplicates were analyzed for each sex. MpEF1α was used as a control.


- A
A gametangiophore of MpEF1αpro:SUF/suf‐30 ge [Y], indicating the inability of transgenic SUF overexpression in rescuing the feminization phenotype of suf ge [Y]. Scale bar, 2 mm.
- B
Real‐time RT–PCR analyses confirming SUF transcript accumulation in SUF‐overexpressing lines. Constitutively expressed MpEF1α was used as a control. Measurements of six biological replicates for WT [Y] and three biological replicates for each SUF‐overexpressing line are plotted.
- C
Structures of the gMpFGMYB‐S and gMpFGMYB‐L transgenes without (−S) or with (−L) the putative promoter and the TSS of SUF.
- D
Real‐time RT–PCR measurement of MpFGMYB transcript levels. Three biological replicates are analyzed for each line.
- E–H
Gametangiophores (E, G) and gametangia (F, H) of gMpFGMYB‐S [Y] (E, F) and gMpFGMYB‐L [Y] (G, H). Scale bars, 1 mm (E, G), 50 μm (F), 100 μm (H).
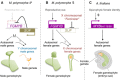
- A, B
A bidirectional transcription module at the MpFGMYB/SUF locus acts as a toggle switch between male (A) and female (B) sexual differentiation in M. polymorpha. MpFGMYB expression is activated by one or more unknown cues associated with reproductive growth. In males, MpFGMYB expression is suppressed by constitutively expressed antisense gene, SUF, allowing an unknown factor (M) to activate both autosomal and Y chromosomal genes to promote male differentiation (A). A dominant “Feminizer” on the X chromosome (Haupt, 1932; Bowman et al, 2017) directly or indirectly suppresses SUF expression, allowing expression of MpFGMYB and downstream autosomal and X chromosomal genes to promote female differentiation (B). Genes on the X and Y chromosomes are dispensable for the sexual morphologies of the gametophytes, but required for the differentiation of functional gametes.
- C
In A. thaliana, three FGMYB genes promote female differentiation in the embryo sac, a highly reduced female gametophyte of flowering plants. FGMYB expression is regulated at the transcriptional level after the formation of sporophytic female floral organs (Kasahara et al, 2005; Rabiger & Drews, 2013).
Comment in
-
An ancient antisense-driven RNA switch drives plant sex determination.EMBO J. 2019 Mar 15;38(6):e101685. doi: 10.15252/embj.2019101685. Epub 2019 Feb 26. EMBO J. 2019. PMID: 30808644 Free PMC article.
Similar articles
-
Transcription of the Antisense Long Non-Coding RNA, SUPPRESSOR OF FEMINIZATION, Represses Expression of the Female-Promoting Gene FEMALE GAMETOPHYTE MYB in the Liverwort Marchantia polymorpha.Plant Cell Physiol. 2024 Apr 16;65(3):338-349. doi: 10.1093/pcp/pcad170. Plant Cell Physiol. 2024. PMID: 38174428 Free PMC article.
-
Three-Dimensional Morphological Analysis Revealed the Cell Patterning Bases for the Sexual Dimorphism Development in the Liverwort Marchantia polymorpha.Plant Cell Physiol. 2023 Aug 17;64(8):866-879. doi: 10.1093/pcp/pcad048. Plant Cell Physiol. 2023. PMID: 37225421
-
Building new insights in plant gametogenesis from an evolutionary perspective.Nat Plants. 2019 Jul;5(7):663-669. doi: 10.1038/s41477-019-0466-0. Epub 2019 Jul 8. Nat Plants. 2019. PMID: 31285561 Review.
-
A bHLH heterodimer regulates germ cell differentiation in land plant gametophytes.Curr Biol. 2023 Nov 20;33(22):4980-4987.e6. doi: 10.1016/j.cub.2023.09.020. Epub 2023 Sep 29. Curr Biol. 2023. PMID: 37776860
-
Meiotic and mitotic cell cycle mutants involved in gametophyte development in Arabidopsis.Mol Plant. 2008 Jul;1(4):564-74. doi: 10.1093/mp/ssn033. Mol Plant. 2008. PMID: 19825562 Review.
Cited by
-
Transcription of the Antisense Long Non-Coding RNA, SUPPRESSOR OF FEMINIZATION, Represses Expression of the Female-Promoting Gene FEMALE GAMETOPHYTE MYB in the Liverwort Marchantia polymorpha.Plant Cell Physiol. 2024 Apr 16;65(3):338-349. doi: 10.1093/pcp/pcad170. Plant Cell Physiol. 2024. PMID: 38174428 Free PMC article.
-
Far-Red Light-Induced Azolla filiculoides Symbiosis Sexual Reproduction: Responsive Transcripts of Symbiont Nostoc azollae Encode Transporters Whilst Those of the Fern Relate to the Angiosperm Floral Transition.Front Plant Sci. 2021 Aug 11;12:693039. doi: 10.3389/fpls.2021.693039. eCollection 2021. Front Plant Sci. 2021. PMID: 34456937 Free PMC article.
-
LncRNAs exert indispensable roles in orchestrating the interaction among diverse noncoding RNAs and enrich the regulatory network of plant growth and its adaptive environmental stress response.Hortic Res. 2023 Nov 17;10(12):uhad234. doi: 10.1093/hr/uhad234. eCollection 2023 Dec. Hortic Res. 2023. PMID: 38156284 Free PMC article.
-
Gamete expression of TALE class HD genes activates the diploid sporophyte program in Marchantia polymorpha.Elife. 2021 Sep 17;10:e57088. doi: 10.7554/eLife.57088. Elife. 2021. PMID: 34533136 Free PMC article.
-
Identification of the sex-determining factor in the liverwort Marchantia polymorpha reveals unique evolution of sex chromosomes in a haploid system.Curr Biol. 2021 Dec 20;31(24):5522-5532.e7. doi: 10.1016/j.cub.2021.10.023. Epub 2021 Nov 3. Curr Biol. 2021. PMID: 34735792 Free PMC article.
References
-
- Althoff F, Kopischke S, Zobell O, Ide K, Ishizaki K, Kohchi T, Zachgo S (2014) Comparison of the MpEF1alpha and CaMV35 promoters for application in Marchantia polymorpha overexpression studies. Transgenic Res 23: 235–244 - PubMed
-
- Bowman JL, Smyth DR, Meyerowitz EM (1991) Genetic interactions among floral homeotic genes of Arabidopsis . Development 112: 1–20 - PubMed
-
- Bowman JL (2016) A brief history of Marchantia from Greece to genomics. Plant Cell Physiol 57: 210–229 - PubMed
-
- Bowman JL, Araki T, Arteaga‐Vazquez MA, Berger F, Dolan L, Haseloff J, Ishizaki K, Kyozuka J, Lin SS, Nagasaki H, Nakagami H, Nakajima K, Nakamura Y, Ohashi‐Ito K, Sawa S, Shimamura M, Solano R, Tsukaya H, Ueda T, Watanabe Y et al (2016) The naming of names: guidelines for gene nomenclature in Marchantia . Plant Cell Physiol 57: 257–261 - PMC - PubMed

