Binding of the Magnaporthe oryzae Chitinase MoChia1 by a Rice Tetratricopeptide Repeat Protein Allows Free Chitin to Trigger Immune Responses
- PMID: 30610168
- PMCID: PMC6391695
- DOI: 10.1105/tpc.18.00382
Binding of the Magnaporthe oryzae Chitinase MoChia1 by a Rice Tetratricopeptide Repeat Protein Allows Free Chitin to Trigger Immune Responses
Abstract
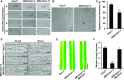
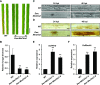
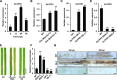
To defend against pathogens, plants have developed complex immune systems, including plasma membrane receptors that recognize pathogen-associated molecular patterns, such as chitin from fungal cell walls, and mount a defense response. Here, we identify a chitinase, MoChia1 (Magnaporthe oryzae chitinase 1), secreted by M. oryzae, a fungal pathogen of rice (Oryza sativa). MoChia1 can trigger plant defense responses, and expression of MoChia1 under an inducible promoter in rice enhances its resistance to M. oryzae MoChia1 is a functional chitinase required for M. oryzae growth and development; knocking out MoChia1 significantly reduced the virulence of the fungus, and we found that MoChia1 binds chitin to suppress the chitin-triggered plant immune response. However, the rice tetratricopeptide repeat protein OsTPR1 interacts with MoChia1 in the rice apoplast. OsTPR1 competitively binds MoChia1, thereby allowing the accumulation of free chitin and re-establishing the immune response. Overexpressing OsTPR1 in rice plants resulted in elevated levels of reactive oxygen species during M. oryzae infection. Our data demonstrate that rice plants not only recognize MoChia1, but also use OsTPR to counteract the function of this fungal chitinase and regain immunity.
© 2019 American Society of Plant Biologists. All rights reserved.
Figures






Similar articles
-
Effector-mediated suppression of chitin-triggered immunity by magnaporthe oryzae is necessary for rice blast disease.Plant Cell. 2012 Jan;24(1):322-35. doi: 10.1105/tpc.111.092957. Epub 2012 Jan 20. Plant Cell. 2012. PMID: 22267486 Free PMC article.
-
A Magnaporthe Chitinase Interacts with a Rice Jacalin-Related Lectin to Promote Host Colonization.Plant Physiol. 2019 Apr;179(4):1416-1430. doi: 10.1104/pp.18.01594. Epub 2019 Jan 29. Plant Physiol. 2019. PMID: 30696749 Free PMC article.
-
Toward understanding of rice innate immunity against Magnaporthe oryzae.Crit Rev Biotechnol. 2016;36(1):165-74. doi: 10.3109/07388551.2014.946883. Epub 2014 Sep 8. Crit Rev Biotechnol. 2016. PMID: 25198435 Review.
-
The Magnaporthe oryzae effector AvrPiz-t targets the RING E3 ubiquitin ligase APIP6 to suppress pathogen-associated molecular pattern-triggered immunity in rice.Plant Cell. 2012 Nov;24(11):4748-62. doi: 10.1105/tpc.112.105429. Epub 2012 Nov 30. Plant Cell. 2012. PMID: 23204406 Free PMC article.
-
Current understanding of pattern-triggered immunity and hormone-mediated defense in rice (Oryza sativa) in response to Magnaporthe oryzae infection.Semin Cell Dev Biol. 2018 Nov;83:95-105. doi: 10.1016/j.semcdb.2017.10.020. Epub 2017 Nov 2. Semin Cell Dev Biol. 2018. PMID: 29061483 Review.
Cited by
-
The Cysteine-Rich Repeat Protein TaCRR1 Participates in Defense against Both Rhizoctonia cerealis and Bipolaris sorokiniana in Wheat.Int J Mol Sci. 2020 Aug 9;21(16):5698. doi: 10.3390/ijms21165698. Int J Mol Sci. 2020. PMID: 32784820 Free PMC article.
-
Understanding the Rice Fungal Pathogen Tilletia horrida from Multiple Perspectives.Rice (N Y). 2022 Dec 16;15(1):64. doi: 10.1186/s12284-022-00612-1. Rice (N Y). 2022. PMID: 36522490 Free PMC article. Review.
-
Fungal effectors, the double edge sword of phytopathogens.Curr Genet. 2021 Feb;67(1):27-40. doi: 10.1007/s00294-020-01118-3. Epub 2020 Nov 4. Curr Genet. 2021. PMID: 33146780 Review.
-
Understanding the Dynamics of Blast Resistance in Rice-Magnaporthe oryzae Interactions.J Fungi (Basel). 2022 May 30;8(6):584. doi: 10.3390/jof8060584. J Fungi (Basel). 2022. PMID: 35736067 Free PMC article. Review.
-
Comparative profiling of canonical and non-canonical small RNAs in the rice blast fungus, Magnaporthe oryzae.Front Microbiol. 2022 Sep 26;13:995334. doi: 10.3389/fmicb.2022.995334. eCollection 2022. Front Microbiol. 2022. PMID: 36225371 Free PMC article.
References
-
- Boutrot F., Zipfel C. (2017). Function, discovery, and exploitation of plant pattern recognition receptors for broad-spectrum disease resistance. Annu. Rev. Phytopathol. 55: 257–286. - PubMed
-
- Cao J., Yang C., Li L., Jiang L., Wu Y., Wu C., Bu Q., Xia G., Liu X., Luo Y., Liu J. (2016). Rice Plasma membrane proteomics reveals Magnaporthe oryzae promotes susceptibility by sequential activation of host hormone signaling pathways. Mol. Plant Microbe Interact. 29: 902–913. - PubMed
-
- Cao J., Yu Y., Huang J., Liu R., Chen Y., Li S., Liu J. (2017). Genome re-sequencing analysis uncovers pathogenecity-related genes undergoing positive selection in Magnaporthe oryzae. Sci. China Life Sci. 60: 880–890. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

