The Phosphoinositide Kinase PIKfyve Promotes Cathepsin-S-Mediated Major Histocompatibility Complex Class II Antigen Presentation
- PMID: 30612035
- PMCID: PMC6319320
- DOI: 10.1016/j.isci.2018.12.015
The Phosphoinositide Kinase PIKfyve Promotes Cathepsin-S-Mediated Major Histocompatibility Complex Class II Antigen Presentation
Abstract
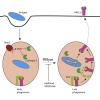
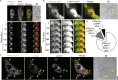
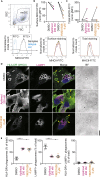
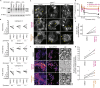
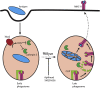
Antigen presentation to T cells in major histocompatibility complex class II (MHC class II) requires the conversion of early endo/phagosomes into lysosomes by a process called maturation. Maturation is driven by the phosphoinositide kinase PIKfyve. Blocking PIKfyve activity by small molecule inhibitors caused a delay in the conversion of phagosomes into lysosomes and in phagosomal acidification, whereas production of reactive oxygen species (ROS) increased. Elevated ROS resulted in reduced activity of cathepsin S and B, but not X, causing a proteolytic defect of MHC class II chaperone invariant chain Ii processing. We developed a novel universal MHC class II presentation assay based on a bio-orthogonal "clickable" antigen and showed that MHC class II presentation was disrupted by the inhibition of PIKfyve, which in turn resulted in reduced activation of CD4+ T cells. Our results demonstrate a key role of PIKfyve in the processing and presentation of antigens, which should be taken into consideration when targeting PIKfyve in autoimmune disease and cancer.
Keywords: Immune Response; Immunology; Molecular Mechanism of Gene Regulation.
Copyright © 2018 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures



References
-
- Allan E.R., Tailor P., Balce D.R., Pirzadeh P., McKenna N.T., Renaux B., Warren A.L., Jirik F.R., Yates R.M. NADPH oxidase modifies patterns of MHC class II-restricted epitopic repertoires through redox control of antigen processing. J. Immunol. 2014;192:4989–5001. - PubMed
-
- Amigorena S., Drake J.R., Webster P., Mellman I. Transient accumulation of new class II MHC molecules in a novel endocytic compartment in B lymphocytes. Nature. 1994;369:113–120. - PubMed
-
- Anderson K.E., Chessa T.A., Davidson K., Henderson R.B., Walker S., Tolmachova T., Grys K., Rausch O., Seabra M.C., Tybulewicz V.L. PtdIns3P and Rac direct the assembly of the NADPH oxidase on a novel, pre-phagosomal compartment during FcR-mediated phagocytosis in primary mouse neutrophils. Blood. 2010;116:4978–4989. - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

