Late Endosomes Act as mRNA Translation Platforms and Sustain Mitochondria in Axons
- PMID: 30612743
- PMCID: PMC6333918
- DOI: 10.1016/j.cell.2018.11.030
Late Endosomes Act as mRNA Translation Platforms and Sustain Mitochondria in Axons
Abstract
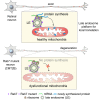
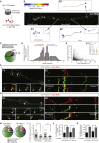
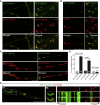
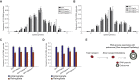
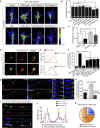
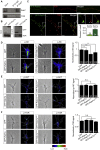
Local translation regulates the axonal proteome, playing an important role in neuronal wiring and axon maintenance. How axonal mRNAs are localized to specific subcellular sites for translation, however, is not understood. Here we report that RNA granules associate with endosomes along the axons of retinal ganglion cells. RNA-bearing Rab7a late endosomes also associate with ribosomes, and real-time translation imaging reveals that they are sites of local protein synthesis. We show that RNA-bearing late endosomes often pause on mitochondria and that mRNAs encoding proteins for mitochondrial function are translated on Rab7a endosomes. Disruption of Rab7a function with Rab7a mutants, including those associated with Charcot-Marie-Tooth type 2B neuropathy, markedly decreases axonal protein synthesis, impairs mitochondrial function, and compromises axonal viability. Our findings thus reveal that late endosomes interact with RNA granules, translation machinery, and mitochondria and suggest that they serve as sites for regulating the supply of nascent pro-survival proteins in axons.
Keywords: Charcot-Marie-Tooth type 2B; RNA trafficking; axon maintenance; endosome; local protein synthesis; lysosome; mRNA translation; mitochondria; retinal ganglion cell.
Copyright © 2018 The Authors. Published by Elsevier Inc. All rights reserved.
Figures







References
-
- Andreassi C., Riccio A. To localize or not to localize: mRNA fate is in 3’UTR ends. Trends Cell Biol. 2009;19:465–474. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 203249/Z/16/Z/WT_/Wellcome Trust/United Kingdom
- 322817/ERC_/European Research Council/International
- 100329/Z/12/Z/WT_/Wellcome Trust/United Kingdom
- BB/N006402/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/N006402/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

