Computational protein structure refinement: Almost there, yet still so far to go
- PMID: 30613211
- PMCID: PMC6319934
- DOI: 10.1002/wcms.1307
Computational protein structure refinement: Almost there, yet still so far to go
Abstract
Protein structures are essential in modern biology yet experimental methods are far from being able to catch up with the rapid increase in available genomic data. Computational protein structure prediction methods aim to fill the gap while the role of protein structure refinement is to take approximate initial template-based models and bring them closer to the true native structure. Current methods for computational structure refinement rely on molecular dynamics simulations, related sampling methods, or iterative structure optimization protocols. The best methods are able to achieve moderate degrees of refinement but consistent refinement that can reach near-experimental accuracy remains elusive. Key issues revolve around the accuracy of the energy function, the inability to reliably rank multiple models, and the use of restraints that keep sampling close to the native state but also limit the degree of possible refinement. A different aspect is the question of what exactly the target of high-resolution refinement should be as experimental structures are affected by experimental conditions and different biological questions require varying levels of accuracy. While improvement of the global protein structure is a difficult problem, high-resolution refinement methods that improves local structural quality such as favorable stereochemistry and the avoidance of atomic clashes are much more successful.
Conflict of interest statement
The author declares no competing interests.
Figures
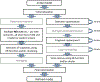
References
-
- Perutz MF, Rossmann MG, Cullis AF, Muirhead H, Will G, North ACT. Structure of Haemoglobin - 3-dimensional Fourier Synthesis at 5.5-Å Resolution, Obtained by X-ray Analysis. Nature 1960, 185:416–422. - PubMed
-
- Kendrew JC, Bodo G, Dintzis HM, Parrish RG, Wyckoff H, Phillips DC. 3-dimensional Model of the Myoglobin Molecule Obtained by X-ray Analysis. Nature 1958, 181:662–666. - PubMed
-
- Whisstock JC, Lesk AM. Prediction of protein function from protein sequence and structure. Q. Rev. Biophys. 2003, 36:307–340. - PubMed
-
- Gane PJ, Dean PM. Recent Advances in structure-based rational drug design. Curr. Opin. Struct. Biol. 2000, 10:401–404. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
