Circumventing senescence is associated with stem cell properties and metformin sensitivity
- PMID: 30614183
- PMCID: PMC6413657
- DOI: 10.1111/acel.12889
Circumventing senescence is associated with stem cell properties and metformin sensitivity
Abstract
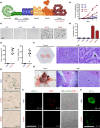
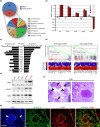
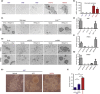
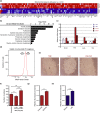
Most cancers arise in old individuals, which also accumulate senescent cells. Cellular senescence can be experimentally induced by expression of oncogenes or telomere shortening during serial passage in culture. In vivo, precursor lesions of several cancer types accumulate senescent cells, which are thought to represent a barrier to malignant progression and a response to the aberrant activation of growth signaling pathways by oncogenes (oncogene toxicity). Here, we sought to define gene expression changes associated with cells that bypass senescence induced by oncogenic RAS. In the context of pancreatic ductal adenocarcinoma (PDAC), oncogenic KRAS induces benign pancreatic intraepithelial neoplasias (PanINs), which exhibit features of oncogene-induced senescence. We found that the bypass of senescence in PanINs leads to malignant PDAC cells characterized by gene signatures of epithelial-mesenchymal transition, stem cells, and mitochondria. Stem cell properties were similarly acquired in PanIN cells treated with LPS, and in primary fibroblasts and mammary epithelial cells that bypassed Ras-induced senescence after reduction of ERK signaling. Intriguingly, maintenance of cells that circumvented senescence and acquired stem cell properties was blocked by metformin, an inhibitor of complex I of the electron transport chain or depletion of STAT3, a protein required for mitochondrial functions and stemness. Thus, our studies link bypass of senescence in premalignant lesions to loss of differentiation, acquisition of stemness features, and increased reliance on mitochondrial functions.
© 2019 The Authors. Aging Cell published by the Anatomical Society and John Wiley & Sons Ltd.
Conflict of interest statement
None declared.
Figures


References
-
- Campbell, K. J. , Rocha, S. , & Perkins, N. D. (2004). Active repression of antiapoptotic gene expression by RelA(p65) NF‐kappa B. Molecular Cell, 13(6), 853–865. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

