Proteomic characterization and discrimination of Aeromonas species recovered from meat and water samples with a spotlight on the antimicrobial resistance of Aeromonas hydrophila
- PMID: 30614207
- PMCID: PMC6854848
- DOI: 10.1002/mbo3.782
Proteomic characterization and discrimination of Aeromonas species recovered from meat and water samples with a spotlight on the antimicrobial resistance of Aeromonas hydrophila
Abstract
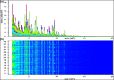
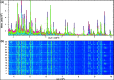
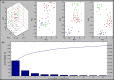
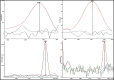
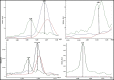
Aeromonas is recognized as a human pathogen following ingestion of contaminated food and water. One major problem in Aeromonas identification is that certain species are phenotypically very similar. The antimicrobial resistance is another significant challenge worldwide. We therefore aimed to use mass spectrometry technology for identification and discrimination of Aeromonas species and to screen the antimicrobial resistance of Aeromonas hydrophila (A. hydrophila). A total of 150 chicken meat and water samples were cultured, and then, the isolates were identified biochemically by the Vitek® 2 Compact system. Proteomic identification was performed by MALDI-TOF MS and confirmed by a microchannel fluidics electrophoresis assay. Principal component analysis (PCA) and single-peak analysis created by MALDI were also used to discriminate the Aeromonas species. The antimicrobial resistance of the A. hydrophila isolates was determined by Vitek® 2 AST cards. In total, 43 samples were positive for Aeromonas and comprised 22 A. hydrophila, 12 Aeromonas caviae (A. caviae), and 9 Aeromonas sobria (A. sobria) isolates. Thirty-nine out of 43 (90.69%) Aeromonas isolates were identified by the Vitek® 2 Compact system, whereas 100% of the Aeromonas isolates were correctly identified by MALDI-TOF MS with a score value ≥2.00. PCA successfully separated A. hydrophila, A. caviae and A. sobria isolates into two groups. Single-peak analysis revealed four discriminating peaks that separated A. hydrophila from A. caviae and A. sobria isolates. The resistance of A. hydrophila to antibiotics was 95.46% for ampicillin, 50% for cefotaxime, 45.45% for norfloxacin and pefloxacin, 36.36% for ceftazidime and ciprofloxacin, 31.81% for ofloxacin and 27.27% for nalidixic acid and tobramycin. In conclusion, chicken meat and water were tainted with Aeromonas spp., with a high occurrence of A. hydrophila. MALDI-TOF MS is a powerful technique for characterizing aeromonads at the genus and species levels. Future studies should investigate the resistance of A. hydrophila to various antimicrobial agents.
Keywords: Aeromonas spp.; antimicrobial resistance; differentiation; microchannel electrophoresis; protein fingerprinting.
© 2019 The Authors. MicrobiologyOpen published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
References
-
- Aboyadak, I. M. , Ali, N. G. , Goda, A. M. , Saad, W. , & Salam, A. M. E. (2017). Non‐selectivity of R‐S media for Aeromonas hydrophila and TCBS media for Vibrio species isolated from Diseased Oreochromis niloticus . Journal of Aquaculture Research and Development, 8, 496–500. 10.4172/2155-9546.1000496 - DOI
-
- Adebayo, E. A. , Majolagbe, O. N. , Ola, I. O. , & Ogundiran, M. A. (2012). Antibiotic resistance pattern of isolated bacterial from salads. Journal of Research in Biology, 2, 136–142.
-
- Ahmed, N. I. , Abd El Aal, S. F. A. , Ayoub, M. A. , & El Sayed, M. S. (2014). Enumeration and characterization of Aeromonas spp. isolated from milk and some dairy products in Sharkia Governorate Egypt. Alexandria Journal of Veterinary Sciences, 40, 52–64.
Publication types
MeSH terms
Substances
Supplementary concepts
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

