Elucidating synergistic dependencies in lung adenocarcinoma by proteome-wide signaling-network analysis
- PMID: 30615629
- PMCID: PMC6322741
- DOI: 10.1371/journal.pone.0208646
Elucidating synergistic dependencies in lung adenocarcinoma by proteome-wide signaling-network analysis
Abstract
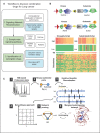
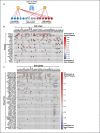
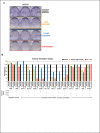
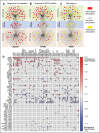
To understand drug combination effect, it is necessary to decipher the interactions between drug targets-many of which are signaling molecules. Previously, such signaling pathway models are largely based on the compilation of literature data from heterogeneous cellular contexts. Indeed, de novo reconstruction of signaling interactions from large-scale molecular profiling is still lagging, compared to similar efforts in transcriptional and protein-protein interaction networks. To address this challenge, we introduce a novel algorithm for the systematic inference of protein kinase pathways, and applied it to published mass spectrometry-based phosphotyrosine profile data from 250 lung adenocarcinoma (LUAD) samples. The resulting network includes 43 TKs and 415 inferred, LUAD-specific substrates, which were validated at >60% accuracy by SILAC assays, including "novel' substrates of the EGFR and c-MET TKs, which play a critical oncogenic role in lung cancer. This systematic, data-driven model supported drug response prediction on an individual sample basis, including accurate prediction and validation of synergistic EGFR and c-MET inhibitor activity in cells lacking mutations in either gene, thus contributing to current precision oncology efforts.
Conflict of interest statement
AC is founder, equity holder, consultant, and director of DarwinHealth Inc., a company that has licensed some of the algorithms used in this manuscript from Columbia University. Columbia University is also an equity holder in DarwinHealth Inc. The ARACNe and MARINa algorithms discussed in this manuscript are publicly and freely available to any researchers working for a non-profit/academic institution but their commercial use is restricted since they were exclusively licensed by Columbia University to DarwinHealth Inc. This does not alter our adherence to PLOS ONE policies on sharing data and materials.
Figures


References
-
- Ettinger DS, Akerley W, Borghaei H, Chang AC, Cheney RT, Chirieac LR, et al. Non-small cell lung cancer. Journal of the National Comprehensive Cancer Network: JNCCN. 2012;1s(10):1236–71. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

