Genome-wide dynamic network analysis reveals a critical transition state of flower development in Arabidopsis
- PMID: 30616516
- PMCID: PMC6323737
- DOI: 10.1186/s12870-018-1589-6
Genome-wide dynamic network analysis reveals a critical transition state of flower development in Arabidopsis
Abstract
Background: The flowering transition which is controlled by a complex and intricate gene regulatory network plays an important role in the reproduction for offspring of plants. It is a challenge to identify the critical transition state as well as the genes that control the transition of flower development. With the emergence of massively parallel sequencing, a great number of time-course transcriptome data greatly facilitate the exploration of the developmental phase transition in plants. Although some network-based bioinformatics analyses attempted to identify the genes that control the phase transition, they generally overlooked the dynamics of regulation and resulted in unreliable results. In addition, the results of these methods cannot be self-explained.
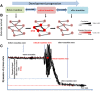
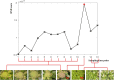
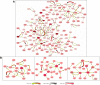
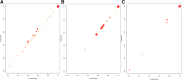
Results: In this work, to reveal a critical transition state and identify the transition-specific genes of flower development, we implemented a genome-wide dynamic network analysis on temporal gene expression data in Arabidopsis by dynamic network biomarker (DNB) method. In the analysis, DNB model which can exploit collective fluctuations and correlations of different metabolites at a network level was used to detect the imminent critical transition state or the tipping point. The genes that control the phase transition can be identified by the difference of weighted correlations between the genes interested and the other genes in the global network. To construct the gene regulatory network controlling the flowering transition, we applied NARROMI algorithm which can reduce the noisy, redundant and indirect regulations on the expression data of the transition-specific genes. In the results, the critical transition state detected during the formation of flowers corresponded to the development of flowering on the 7th to 9th day in Arabidopsis. Among of 233 genes identified to be highly fluctuated at the transition state, a high percentage of genes with maximum expression in pollen was detected, and 24 genes were validated to participate in stress reaction process, as well as other floral-related pathways. Composed of three major subnetworks, a gene regulatory network with 150 nodes and 225 edges was found to be highly correlated with flowering transition. The gene ontology (GO) annotation of pathway enrichment analysis revealed that the identified genes are enriched in the catalytic activity, metabolic process and cellular process.
Conclusions: This study provides a novel insight to identify the real causality of the phase transition with genome-wide dynamic network analysis.
Keywords: Dynamical network biomarker (DNB); Flower development; Gene regulatory network (GRN); Phase transition; Time-course gene expression data.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

