Molecular Mechanisms of Macular Degeneration Associated with the Complement Factor H Y402H Mutation
- PMID: 30616835
- PMCID: PMC6350009
- DOI: 10.1016/j.bpj.2018.12.007
Molecular Mechanisms of Macular Degeneration Associated with the Complement Factor H Y402H Mutation
Abstract
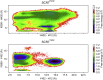
A single nucleotide polymorphism, tyrosine at position 402 to histidine (Y402H), within the gene encoding complement factor H (FH) predisposes individuals to acquiring age-related macular degeneration (AMD) after aging. This polymorphism occurs in short consensus repeat (SCR) 7 of FH and results in decreased binding affinity of SCR6-8 for heparin. As FH is responsible for regulating the complement system, decreased affinity for heparin results in decreased regulation on surfaces of self. To understand the involvement of the Y402H polymorphism in AMD, we leverage methods from bioinformatics and computational biophysics to quantify structural and dynamical differences between SCR7 isoforms that contribute to decreased pattern recognition in SCR7H402. Our data from molecular and Brownian dynamics simulations suggest a revised mechanism for decreased heparin binding. In this model, transient contacts not observed in structures for SCR7 are predicted to occur in molecular dynamics simulations between coevolved residues Y402 and I412, stabilizing SCR7Y402 in a conformation that promotes association with heparin. H402 in the risk isoform is less likely to form a contact with I412 and samples a larger conformational space than Y402. We observe energy minima for sidechains of Y402 and R404 from SCR7Y402 that are predicted to associate with heparin at a rate constant faster than energy minima for sidechains of H402 and R404 from SCR7H402. As both carbohydrate density and degree of sulfation decrease with age in Bruch's membrane of the macula, the decreased heparin recognition of SCR7H402 may contribute to the pathogenesis of AMD.
Copyright © 2018 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures
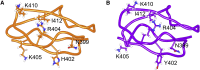






References
-
- de Juan D., Pazos F., Valencia A. Emerging methods in protein co-evolution. Nat. Rev. Genet. 2013;14:249–261. - PubMed
-
- Liszewski M.K., Atkinson J.P. Complement regulators in human disease: lessons from modern genetics. J. Intern. Med. 2015;277:294–305. - PubMed
-
- Herbert A.P., Deakin J.A., Barlow P.N. Structure shows that a glycosaminoglycan and protein recognition site in factor H is perturbed by age-related macular degeneration-linked single nucleotide polymorphism. J. Biol. Chem. 2007;282:18960–18968. - PubMed
-
- Clark S.J., Higman V.A., Day A.J. His-384 allotypic variant of factor H associated with age-related macular degeneration has different heparin binding properties from the non-disease-associated form. J. Biol. Chem. 2006;281:24713–24720. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

