GADD45A binds R-loops and recruits TET1 to CpG island promoters
- PMID: 30617255
- PMCID: PMC6420098
- DOI: 10.1038/s41588-018-0306-6
GADD45A binds R-loops and recruits TET1 to CpG island promoters
Abstract
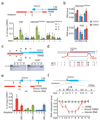
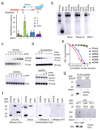
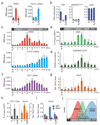
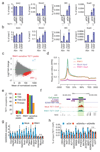
R-loops are DNA-RNA hybrids enriched at CpG islands (CGIs) that can regulate chromatin states1-8. How R-loops are recognized and interpreted by specific epigenetic readers is unknown. Here we show that GADD45A (growth arrest and DNA damage protein 45A) binds directly to R-loops and mediates local DNA demethylation by recruiting TET1 (ten-eleven translocation 1). Studying the tumor suppressor TCF21 (ref. 9), we find that antisense long noncoding (lncRNA) TARID (TCF21 antisense RNA inducing promoter demethylation) forms an R-loop at the TCF21 promoter. Binding of GADD45A to the R-loop triggers local DNA demethylation and TCF21 expression. TARID transcription, R-loop formation, DNA demethylation, and TCF21 expression proceed sequentially during the cell cycle. Oxidized DNA demethylation intermediates are enriched at genomic R-loops and their levels increase upon RNase H1 depletion. Genomic profiling in embryonic stem cells identifies thousands of R-loop-dependent TET1 binding sites at CGIs. We propose that GADD45A is an epigenetic R-loop reader that recruits the demethylation machinery to promoter CGIs.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

