Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer's disease risk
- PMID: 30617256
- PMCID: PMC6836675
- DOI: 10.1038/s41588-018-0311-9
Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer's disease risk
Erratum in
-
Author Correction: Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer's disease risk.Nat Genet. 2020 Mar;52(3):354. doi: 10.1038/s41588-019-0573-x. Nat Genet. 2020. PMID: 32029921
Abstract
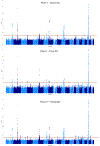
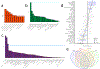
Alzheimer's disease (AD) is highly heritable and recent studies have identified over 20 disease-associated genomic loci. Yet these only explain a small proportion of the genetic variance, indicating that undiscovered loci remain. Here, we performed a large genome-wide association study of clinically diagnosed AD and AD-by-proxy (71,880 cases, 383,378 controls). AD-by-proxy, based on parental diagnoses, showed strong genetic correlation with AD (rg = 0.81). Meta-analysis identified 29 risk loci, implicating 215 potential causative genes. Associated genes are strongly expressed in immune-related tissues and cell types (spleen, liver, and microglia). Gene-set analyses indicate biological mechanisms involved in lipid-related processes and degradation of amyloid precursor proteins. We show strong genetic correlations with multiple health-related outcomes, and Mendelian randomization results suggest a protective effect of cognitive ability on AD risk. These results are a step forward in identifying the genetic factors that contribute to AD risk and add novel insights into the neurobiology of AD.
Conflict of interest statement
Competing Interests Statement
The authors report the following potentially competing financial interests. P.F.S.: Lundbeck (advisory committee), Pfizer (Scientific Advisory Board member), and Roche (grant recipient, speaker reimbursement). J.H.L.: Cartana (Scientific Advisor) and Roche (grant recipient). O.A.A.: Lundbeck (speaker’s honorarium). St.St., H.S., and K.S. are employees of deCODE Genetics/Amgen. J.H. is a cograntee of Cytox from Innovate UK (UK Department of Business). D.A. has received research support and/or honoraria from Astra-Zeneca, Lundbeck, Novartis Pharmaceuticals, and GE Health, and serves as a paid consultant for Lundbeck, Eisai, Heptares, and Axovant. All other authors declare no financial interests or potential conflicts of interest.
Figures

References
-
- Prince M, Bryce R, Albanese E, Wimo A, Ribeiro W, Ferri CP. The global prevalence of dementia: a systematic review and metaanalysis. Alzheimer’s & dementia : the journal of the Alzheimer’s Association 2013; 9(1): 63–75.e2. - PubMed
-
- Gatz M, Reynolds CA, Fratiglioni L, et al. Role of genes and environments for explaining Alzheimer disease. Arch Gen Psychiatry 2006; 63(2): 168–74. - PubMed
-
- Cacace R, Sleegers K, Van Broeckhoven C. Molecular genetics of early-onset Alzheimer’s disease revisited. Alzheimer’s & dementia : the journal of the Alzheimer’s Association 2016; 12(6): 733–48. - PubMed
-
- Goate A, Chartier-Harlin MC, Mullan M, et al. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer’s disease. Nature 1991; 349(6311): 704–6. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

