Enabling genetic analysis of diverse bacteria with Mobile-CRISPRi
- PMID: 30617347
- PMCID: PMC6424567
- DOI: 10.1038/s41564-018-0327-z
Enabling genetic analysis of diverse bacteria with Mobile-CRISPRi
Abstract
The vast majority of bacteria, including human pathogens and microbiome species, lack genetic tools needed to systematically associate genes with phenotypes. This is the major impediment to understanding the fundamental contributions of genes and gene networks to bacterial physiology and human health. Clustered regularly interspaced short palindromic repeats interference (CRISPRi), a versatile method of blocking gene expression using a catalytically inactive Cas9 protein (dCas9) and programmable single guide RNAs, has emerged as a powerful genetic tool to dissect the functions of essential and non-essential genes in species ranging from bacteria to humans1-6. However, the difficulty of establishing effective CRISPRi systems across bacteria is a major barrier to its widespread use to dissect bacterial gene function. Here, we establish 'Mobile-CRISPRi', a suite of CRISPRi systems that combines modularity, stable genomic integration and ease of transfer to diverse bacteria by conjugation. Focusing predominantly on human pathogens associated with antibiotic resistance, we demonstrate the efficacy of Mobile-CRISPRi in gammaproteobacteria and Bacillales Firmicutes at the individual gene scale, by examining drug-gene synergies, and at the library scale, by systematically phenotyping conditionally essential genes involved in amino acid biosynthesis. Mobile-CRISPRi enables genetic dissection of non-model bacteria, facilitating analyses of microbiome function, antibiotic resistances and sensitivities, and comprehensive screens for host-microorganism interactions.
Figures

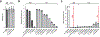
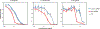

Comment in
-
Harnessing undomesticated life.Nat Microbiol. 2019 Feb;4(2):212-213. doi: 10.1038/s41564-018-0355-8. Nat Microbiol. 2019. PMID: 30675034 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

