The Role of RNA Editing in Cancer Development and Metabolic Disorders
- PMID: 30619092
- PMCID: PMC6305585
- DOI: 10.3389/fendo.2018.00762
The Role of RNA Editing in Cancer Development and Metabolic Disorders
Abstract
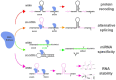
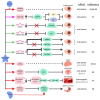
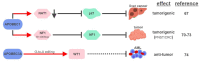
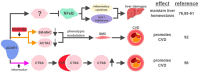
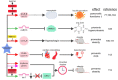
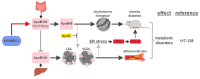
Numerous human diseases arise from alterations of genetic information, most notably DNA mutations. Thought to be merely the intermediate between DNA and protein, changes in RNA sequence were an afterthought until the discovery of RNA editing 30 years ago. RNA editing alters RNA sequence without altering the sequence or integrity of genomic DNA. The most common RNA editing events are A-to-I changes mediated by adenosine deaminase acting on RNA (ADAR), and C-to-U editing mediated by apolipoprotein B mRNA editing enzyme, catalytic polypeptide 1 (APOBEC1). Both A-to-I and C-to-U editing were first identified in the context of embryonic development and physiological homeostasis. The role of RNA editing in human disease has only recently started to be understood. In this review, the impact of RNA editing on the development of cancer and metabolic disorders will be examined. Distinctive functions of each RNA editase that regulate either A-to-I or C-to-U editing will be highlighted in addition to pointing out important regulatory mechanisms governing these processes. The potential of developing novel therapeutic approaches through intervention of RNA editing will be explored. As the role of RNA editing in human disease is elucidated, the clinical utility of RNA editing targeted therapies will be needed. This review aims to serve as a bridge of information between past findings and future directions of RNA editing in the context of cancer and metabolic disease.
Keywords: ADAR; APOBEC1; RNA editing; cancer; metabolic disease.
Figures

References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

