Metabolism and Occurrence of Methanogenic and Sulfate-Reducing Syntrophic Acetate Oxidizing Communities in Haloalkaline Environments
- PMID: 30619130
- PMCID: PMC6295475
- DOI: 10.3389/fmicb.2018.03039
Metabolism and Occurrence of Methanogenic and Sulfate-Reducing Syntrophic Acetate Oxidizing Communities in Haloalkaline Environments
Abstract
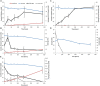
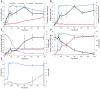
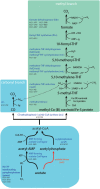
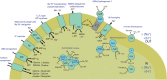
Anaerobic syntrophic acetate oxidation (SAO) is a thermodynamically unfavorable process involving a syntrophic acetate oxidizing bacterium (SAOB) that forms interspecies electron carriers (IECs). These IECs are consumed by syntrophic partners, typically hydrogenotrophic methanogenic archaea or sulfate reducing bacteria. In this work, the metabolism and occurrence of SAOB at extremely haloalkaline conditions were investigated, using highly enriched methanogenic (M-SAO) and sulfate-reducing (S-SAO) cultures from south-western Siberian hypersaline soda lakes. Activity tests with the M-SAO and S-SAO cultures and thermodynamic calculations indicated that H2 and formate are important IECs in both SAO cultures. Metagenomic analysis of the M-SAO cultures showed that the dominant SAOB was 'Candidatus Syntrophonatronum acetioxidans,' and a near-complete draft genome of this SAOB was reconstructed. 'Ca. S. acetioxidans' has all genes necessary for operating the Wood-Ljungdahl pathway, which is likely employed for acetate oxidation. It also encodes several genes essential to thrive at haloalkaline conditions; including a Na+-dependent ATP synthase and marker genes for 'salt-out' strategies for osmotic homeostasis at high soda conditions. Membrane lipid analysis of the M-SAO culture showed the presence of unusual bacterial diether membrane lipids which are presumably beneficial at extreme haloalkaline conditions. To determine the importance of SAO in haloalkaline environments, previously obtained 16S rRNA gene sequencing data and metagenomic data of five different hypersaline soda lake sediment samples were investigated, including the soda lakes where the enrichment cultures originated from. The draft genome of 'Ca. S. acetioxidans' showed highest identity with two metagenome-assembled genomes (MAGs) of putative SAOBs that belonged to the highly abundant and diverse Syntrophomonadaceae family present in the soda lake sediments. The 16S rRNA gene amplicon datasets of the soda lake sediments showed a high similarity of reads to 'Ca. S. acetioxidans' with abundance as high as 1.3% of all reads, whereas aceticlastic methanogens and acetate oxidizing sulfate-reducers were not abundant (≤0.1%) or could not be detected. These combined results indicate that SAO is the primary anaerobic acetate oxidizing pathway at extreme haloalkaline conditions performed by haloalkaliphilic syntrophic consortia.
Keywords: SAOB; genome-centric metagenomics; haloalkaliphiles; soda lakes; syntrophic acetate oxidation; syntrophic acetate oxidizing bacteria; syntrophy.
Figures

Similar articles
-
Syntrophic Acetate-Oxidizing Microbial Consortia Enriched from Full-Scale Mesophilic Food Waste Anaerobic Digesters Showing High Biodiversity and Functional Redundancy.mSystems. 2022 Oct 26;7(5):e0033922. doi: 10.1128/msystems.00339-22. Epub 2022 Sep 8. mSystems. 2022. PMID: 36073802 Free PMC article.
-
Sulfate-dependent acetate oxidation under extremely natron-alkaline conditions by syntrophic associations from hypersaline soda lakes.Microbiology (Reading). 2014 Apr;160(Pt 4):723-732. doi: 10.1099/mic.0.075093-0. Epub 2014 Jan 30. Microbiology (Reading). 2014. PMID: 24482193
-
Syntrophic associations from hypersaline soda lakes converting organic acids and alcohols to methane at extremely haloalkaline conditions.Environ Microbiol. 2016 Sep;18(9):3189-202. doi: 10.1111/1462-2920.13448. Epub 2016 Jul 24. Environ Microbiol. 2016. PMID: 27387660
-
Deep insights into the network of acetate metabolism in anaerobic digestion: focusing on syntrophic acetate oxidation and homoacetogenesis.Water Res. 2021 Feb 15;190:116774. doi: 10.1016/j.watres.2020.116774. Epub 2020 Dec 24. Water Res. 2021. PMID: 33387947 Review.
-
Chemolithotrophic haloalkaliphiles from soda lakes.FEMS Microbiol Ecol. 2005 May 1;52(3):287-95. doi: 10.1016/j.femsec.2005.02.012. FEMS Microbiol Ecol. 2005. PMID: 16329914 Review.
Cited by
-
Alternative Pathways of Acetogenic Ethanol and Methanol Degradation in the Thermophilic Anaerobe Thermacetogenium phaeum.Front Microbiol. 2019 Mar 19;10:423. doi: 10.3389/fmicb.2019.00423. eCollection 2019. Front Microbiol. 2019. PMID: 30949135 Free PMC article.
-
Living in mangroves: a syntrophic scenario unveiling a resourceful microbiome.BMC Microbiol. 2024 Jun 28;24(1):228. doi: 10.1186/s12866-024-03390-6. BMC Microbiol. 2024. PMID: 38943070 Free PMC article.
-
Energy-Conserving Enzyme Systems Active During Syntrophic Acetate Oxidation in the Thermophilic Bacterium Thermacetogenium phaeum.Front Microbiol. 2019 Nov 29;10:2785. doi: 10.3389/fmicb.2019.02785. eCollection 2019. Front Microbiol. 2019. PMID: 31849917 Free PMC article.
-
Genomic Insights into Syntrophic Lifestyle of 'Candidatus Contubernalis alkaliaceticus' Based on the Reversed Wood-Ljungdahl Pathway and Mechanism of Direct Electron Transfer.Life (Basel). 2023 Oct 20;13(10):2084. doi: 10.3390/life13102084. Life (Basel). 2023. PMID: 37895465 Free PMC article.
-
DiSCo: a sequence-based type-specific predictor of Dsr-dependent dissimilatory sulphur metabolism in microbial data.Microb Genom. 2021 Jul;7(7):000603. doi: 10.1099/mgen.0.000603. Microb Genom. 2021. PMID: 34241589 Free PMC article.
References
-
- Balk M., Weijma J., Stams A. J. M. (2002). Thermotoga lettingae sp nov., a novel thermophilic, methanol-degrading bacterium isolated from a thermophilic anaerobic reactor. Int. J. Syst. Evol. Microbiol. 52 1361–1368. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

