AU-Rich Long 3' Untranslated Region Regulates Gene Expression in Bacteria
- PMID: 30619162
- PMCID: PMC6299119
- DOI: 10.3389/fmicb.2018.03080
AU-Rich Long 3' Untranslated Region Regulates Gene Expression in Bacteria
Abstract
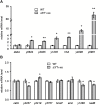
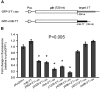
3' untranslated regions (3' UTRs) and particularly long 3' UTRs have been shown to act as a new class of post-transcriptional regulatory element. We previously reported that hmsT mRNA stability is negatively regulated by the 3' UTR of hmsT in Yersinia pestis. To investigate more general effects of 3' UTRs in Y. pestis, we selected 15 genes potentially possessing long 3' UTRs with different AU content and constructed their 3' UTR deletion mutants. Deletion of AU-rich 3' UTRs increased mRNA levels, whereas deletion of 3' UTRs with normal AU content resulted in slight or no changes in the mRNA level. In addition, we found that PNPase was important for 3' UTR-mediated mRNA decay when the transcriptional terminator was Rho-dependent. Finally, we showed that ribosomes promote mRNA stability when bound to a 3' UTR. Our findings suggest that functional 3' UTRs might be broadly distributed in bacteria and their novel regulatory mechanisms require further investigation.
Keywords: 3′ UTR; AU-rich region; Yersinia pestis; mRNA stability; post-transcriptional regulation.
Figures
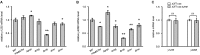
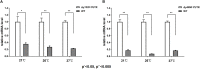
Similar articles
-
Massively parallel analysis of human 3' UTRs reveals that AU-rich element length and registration predict mRNA destabilization.G3 (Bethesda). 2022 Jan 4;12(1):jkab404. doi: 10.1093/g3journal/jkab404. G3 (Bethesda). 2022. PMID: 34849835 Free PMC article.
-
The hmsT 3' untranslated region mediates c-di-GMP metabolism and biofilm formation in Yersinia pestis.Mol Microbiol. 2016 Mar;99(6):1167-78. doi: 10.1111/mmi.13301. Epub 2016 Feb 8. Mol Microbiol. 2016. PMID: 26711808
-
Post-transcriptional regulation of the human inducible nitric oxide synthase (iNOS) expression by the cytosolic poly(A)-binding protein (PABP).Nitric Oxide. 2013 Sep 1;33:6-17. doi: 10.1016/j.niox.2013.05.002. Epub 2013 May 25. Nitric Oxide. 2013. PMID: 23711718
-
Regulation by 3'-Untranslated Regions.Annu Rev Genet. 2017 Nov 27;51:171-194. doi: 10.1146/annurev-genet-120116-024704. Epub 2017 Aug 30. Annu Rev Genet. 2017. PMID: 28853924 Review.
-
Regulatory 3' Untranslated Regions of Bacterial mRNAs.Front Microbiol. 2017 Jul 10;8:1276. doi: 10.3389/fmicb.2017.01276. eCollection 2017. Front Microbiol. 2017. PMID: 28740488 Free PMC article. Review.
Cited by
-
Synthetic Biology Tools for Genome and Transcriptome Engineering of Solventogenic Clostridium.Front Bioeng Biotechnol. 2020 Apr 16;8:282. doi: 10.3389/fbioe.2020.00282. eCollection 2020. Front Bioeng Biotechnol. 2020. PMID: 32363182 Free PMC article. Review.
-
Differential evolution in 3'UTRs leads to specific gene expression in Staphylococcus.Nucleic Acids Res. 2020 Mar 18;48(5):2544-2563. doi: 10.1093/nar/gkaa047. Nucleic Acids Res. 2020. PMID: 32016395 Free PMC article.
-
Regulation of mRNA Stability During Bacterial Stress Responses.Front Microbiol. 2020 Sep 9;11:2111. doi: 10.3389/fmicb.2020.02111. eCollection 2020. Front Microbiol. 2020. PMID: 33013770 Free PMC article. Review.
-
Bacterial 3'UTRs: A Useful Resource in Post-transcriptional Regulation.Front Mol Biosci. 2021 Jan 8;7:617633. doi: 10.3389/fmolb.2020.617633. eCollection 2020. Front Mol Biosci. 2021. PMID: 33490108 Free PMC article. Review.
-
Sensory Perception in Bacterial Cyclic Diguanylate Signal Transduction.J Bacteriol. 2022 Feb 15;204(2):e0043321. doi: 10.1128/JB.00433-21. Epub 2021 Oct 4. J Bacteriol. 2022. PMID: 34606374 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources

