Genomic and in-situ Transcriptomic Characterization of the Candidate Phylum NPL-UPL2 From Highly Alkaline Highly Reducing Serpentinized Groundwater
- PMID: 30619209
- PMCID: PMC6305446
- DOI: 10.3389/fmicb.2018.03141
Genomic and in-situ Transcriptomic Characterization of the Candidate Phylum NPL-UPL2 From Highly Alkaline Highly Reducing Serpentinized Groundwater
Abstract
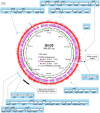
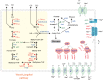
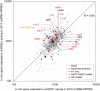
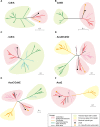
Serpentinization is a process whereby water interacts with reduced mantle rock called peridotite to produce a new suite of minerals (e.g., serpentine), a highly alkaline fluid, and hydrogen. In previous reports, we identified abundance of microbes of the candidate phylum NPL-UPA2 in a serpentinization site called The Cedars. Here, we report the first metagenome assembled genome (MAG) of the candidate phylum as well as the in-situ gene expression. The MAG of the phylum NPL-UPA2, named Unc8, is only about 1 Mbp and its biosynthetic properties suggest it should be capable of independent growth. In keeping with the highly reducing niche of Unc8, its genome encodes none of the known oxidative stress response genes including superoxide dismutases. With regard to energy metabolism, the MAG of Unc8 encodes all enzymes for Wood-Ljungdahl acetogenesis pathway, a ferredoxin:NAD+ oxidoreductase (Rnf) and electron carriers for flavin-based electron bifurcation (Etf, Hdr). Furthermore, the transcriptome of Unc8 in the waters of The Cedars showed enhanced levels of gene expression in the key enzymes of the Wood-Ljungdahl pathway [e.g., Carbon monoxide dehydrogenase /Acetyl-CoA synthase complex (CODH/ACS), Rnf, Acetyl-CoA synthetase (Acd)], which indicated that the Unc8 is an acetogen. However, the MAG of Unc8 encoded no well-known hydrogenase genes, suggesting that the energy metabolism of Unc8 might be focused on CO as the carbon and energy sources for the acetate formation. Given that CO could be supplied via abiotic reaction associated with deep subsurface serpentinization, while available CO2 would be at extremely low concentrations in this high pH environment, CO-associated metabolism could provide advantageous approach. The CODH/ACS in Unc8 is a Bacteria/Archaea hybrid type of six-subunit complex and the electron carriers, Etf and Hdr, showed the highest similarity to those in Archaea, suggesting that archaeal methanogenic energy metabolism was incorporated into the bacterial acetogenesis in NPL-UPA2. Given that serpentinization systems are viewed as potential habitats for early life, and that acetogenesis via the Wood-Ljungdahl pathway is proposed as an energy metabolism of Last Universal Common Ancestor, a phylogenetically distinct acetogen from an early earth analog site may provide important insights in primordial lithotrophs and their habitat.
Keywords: acetogen; alkaliphile ecology; carbon monoxide dehydrogenase; last universal common ancestor; metagenome; metatranscriptome; serpentinization; subsurface microbial community.
Figures



Similar articles
-
Deep-branching acetogens in serpentinized subsurface fluids of Oman.Proc Natl Acad Sci U S A. 2022 Oct 18;119(42):e2206845119. doi: 10.1073/pnas.2206845119. Epub 2022 Oct 10. Proc Natl Acad Sci U S A. 2022. PMID: 36215489 Free PMC article.
-
Genomic Analysis of Calderihabitans maritimus KKC1, a Thermophilic, Hydrogenogenic, Carboxydotrophic Bacterium Isolated from Marine Sediment.Appl Environ Microbiol. 2017 Jul 17;83(15):e00832-17. doi: 10.1128/AEM.00832-17. Print 2017 Aug 1. Appl Environ Microbiol. 2017. PMID: 28526793 Free PMC article.
-
"Candidatus Galacturonibacter soehngenii" Shows Acetogenic Catabolism of Galacturonic Acid but Lacks a Canonical Carbon Monoxide Dehydrogenase/Acetyl-CoA Synthase Complex.Front Microbiol. 2020 Jan 29;11:63. doi: 10.3389/fmicb.2020.00063. eCollection 2020. Front Microbiol. 2020. PMID: 32063897 Free PMC article.
-
Serpentinization: Connecting Geochemistry, Ancient Metabolism and Industrial Hydrogenation.Life (Basel). 2018 Sep 22;8(4):41. doi: 10.3390/life8040041. Life (Basel). 2018. PMID: 30249016 Free PMC article. Review.
-
Energy conservation via electron bifurcating ferredoxin reduction and proton/Na(+) translocating ferredoxin oxidation.Biochim Biophys Acta. 2013 Feb;1827(2):94-113. doi: 10.1016/j.bbabio.2012.07.002. Epub 2012 Jul 16. Biochim Biophys Acta. 2013. PMID: 22800682 Review.
Cited by
-
A non-methanogenic archaeon within the order Methanocellales.Nat Commun. 2024 Jun 13;15(1):4858. doi: 10.1038/s41467-024-48185-5. Nat Commun. 2024. PMID: 38871712 Free PMC article.
-
Microbial ecology of the newly discovered serpentinite-hosted Old City hydrothermal field (southwest Indian ridge).ISME J. 2021 Mar;15(3):818-832. doi: 10.1038/s41396-020-00816-7. Epub 2020 Nov 2. ISME J. 2021. PMID: 33139872 Free PMC article.
-
Deep-branching acetogens in serpentinized subsurface fluids of Oman.Proc Natl Acad Sci U S A. 2022 Oct 18;119(42):e2206845119. doi: 10.1073/pnas.2206845119. Epub 2022 Oct 10. Proc Natl Acad Sci U S A. 2022. PMID: 36215489 Free PMC article.
-
The dynamic history of prokaryotic phyla: discovery, diversity and division.Int J Syst Evol Microbiol. 2024 Sep;74(9):006508. doi: 10.1099/ijsem.0.006508. Int J Syst Evol Microbiol. 2024. PMID: 39250184 Review.
-
Advances in Defining Ecosystem Functions of the Terrestrial Subsurface Biosphere.Front Microbiol. 2022 Jun 2;13:891528. doi: 10.3389/fmicb.2022.891528. eCollection 2022. Front Microbiol. 2022. PMID: 35722320 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

