Butyrate Produced by Commensal Bacteria Down-Regulates Indolamine 2,3-Dioxygenase 1 (IDO-1) Expression via a Dual Mechanism in Human Intestinal Epithelial Cells
- PMID: 30619249
- PMCID: PMC6297836
- DOI: 10.3389/fimmu.2018.02838
Butyrate Produced by Commensal Bacteria Down-Regulates Indolamine 2,3-Dioxygenase 1 (IDO-1) Expression via a Dual Mechanism in Human Intestinal Epithelial Cells
Abstract
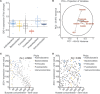
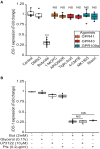
Commensal bacteria are crucial for the development and maintenance of a healthy immune system therefore contributing to the global well-being of their host. A wide variety of metabolites produced by commensal bacteria are influencing host health but the characterization of the multiple molecular mechanisms involved in host-microbiota interactions is still only partially unraveled. The intestinal epithelial cells (IECs) take a central part in the host-microbiota dialogue by inducing the first microbial-derived immune signals. Amongst the numerous effector molecules modulating the immune responses produced by IECs, indoleamine 2,3-dioxygenase-1 (IDO-1) is essential for gut homeostasis. IDO-1 expression is dependent on the microbiota and despites its central role, how the commensal bacteria impacts its expression is still unclear. Therefore, we investigated the impact of individual cultivable commensal bacteria on IDO-1 transcriptional expression and found that the short chain fatty acid (SCFA) butyrate was the main metabolite controlling IDO-1 expression in human primary IECs and IEC cell-lines. This butyrate-driven effect was independent of the G-protein coupled receptors GPR41, GPR43, and GPR109a and of the transcription factors SP1, AP1, and PPARγ for which binding sites were reported in the IDO-1 promoter. We demonstrated for the first time that butyrate represses IDO-1 expression by two distinct mechanisms. Firstly, butyrate decreases STAT1 expression leading to the inhibition of the IFNγ-dependent and phosphoSTAT1-driven transcription of IDO-1. In addition, we described a second mechanism by which butyrate impairs IDO-1 transcription in a STAT1-independent manner that could be attributed to its histone deacetylase (HDAC) inhibitor property. In conclusion, our results showed that IDO-1 expression is down-regulated by butyrate via a dual mechanism: the reduction of STAT1 level and the HDAC inhibitor property of SCFAs.
Keywords: IDO-1; butyrate; gut microbiota; immune gene regulation; intestinal epithelial cells.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

