Spatial Mapping of Myeloid Cells and Macrophages by Multiplexed Tissue Staining
- PMID: 30619287
- PMCID: PMC6302234
- DOI: 10.3389/fimmu.2018.02925
Spatial Mapping of Myeloid Cells and Macrophages by Multiplexed Tissue Staining
Abstract
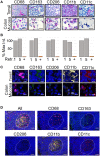
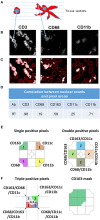
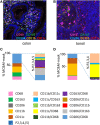
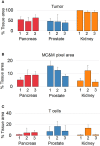
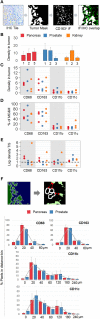
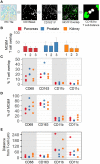
An array of phenotypically diverse myeloid cells and macrophages (MC&M) resides in the tumor microenvironment, requiring multiplexed detection systems for visualization. Here we report an automated, multiplexed staining approach, named PLEXODY, that consists of five MC&M-related fluorescently-tagged antibodies (anti - CD68, - CD163, - CD206, - CD11b, and - CD11c), and three chromogenic antibodies, reactive with high- and low-molecular weight cytokeratins and CD3, highlighting tumor regions, benign glands and T cells. The staining prototype and image analysis methods which include a pixel/area-based quantification were developed using tissues from inflamed colon and tonsil and revealed a unique tissue-specific composition of 14 MC&M-associated pixel classes. As a proof-of-principle, PLEXODY was applied to three cases of pancreatic, prostate and renal cancers. Across digital images from these cancer types we observed 10 MC&M-associated pixel classes at frequencies greater than 3%. Cases revealed higher frequencies of single positive compared to multi-color pixels and a high abundance of CD68+/CD163+ and CD68+/CD163+/CD206+ pixels. Significantly more CD68+ and CD163+ vs. CD11b+ and CD11c+ pixels were in direct contact with tumor cells and T cells. While the greatest percentage (~70%) of CD68+ and CD163+ pixels was 0-20 microns away from tumor and T cell borders, CD11b+ and CD11c+ pixels were detected up to 240 microns away from tumor/T cell masks. Together, these data demonstrate significant differences in densities and spatial organization of MC&M-associated pixel classes, but surprising similarities between the three cancer types.
Keywords: FFPE; immunofluorescence; immunohistochemistry; macrophage; multiplex; myeloid; spatial profiling.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

