A Mass Spectrometry-Based Profiling of Interactomes of Viral DDB1- and Cullin Ubiquitin Ligase-Binding Proteins Reveals NF-κB Inhibitory Activity of the HIV-2-Encoded Vpx
- PMID: 30619335
- PMCID: PMC6305766
- DOI: 10.3389/fimmu.2018.02978
A Mass Spectrometry-Based Profiling of Interactomes of Viral DDB1- and Cullin Ubiquitin Ligase-Binding Proteins Reveals NF-κB Inhibitory Activity of the HIV-2-Encoded Vpx
Abstract
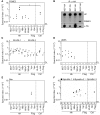
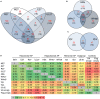
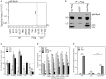
Viruses and hosts are situated in a molecular arms race. To avoid morbidity and mortality, hosts evolved antiviral restriction factors. These restriction factors exert selection pressure on the viruses and drive viral evolution toward increasingly efficient immune antagonists. Numerous viruses exploit cellular DNA damage-binding protein 1 (DDB1)-containing Cullin RocA ubiquitin ligases (CRLs) to induce the ubiquitination and subsequent proteasomal degradation of antiviral factors expressed by their hosts. To establish a comprehensive understanding of the underlying protein interaction networks, we performed immuno-affinity precipitations for a panel of DDB1-interacting proteins derived from viruses such as mouse cytomegalovirus (MCMV, Murid herpesvirus [MuHV] 1), rat cytomegalovirus Maastricht MuHV2, rat cytomegalovirus English MuHV8, human cytomegalovirus (HCMV), hepatitis B virus (HBV), and human immunodeficiency virus (HIV). Cellular interaction partners were identified and quantified by mass spectrometry (MS) and validated by classical biochemistry. The comparative approach enabled us to separate unspecific interactions from specific binding partners and revealed remarkable differences in the strength of interaction with DDB1. Our analysis confirmed several previously described interactions like the interaction of the MCMV-encoded interferon antagonist pM27 with STAT2. We extended known interactions to paralogous proteins like the interaction of the HBV-encoded HBx with different Spindlin proteins and documented interactions for the first time, which explain functional data like the interaction of the HIV-2-encoded Vpr with Bax. Additionally, several novel interactions were identified, such as the association of the HIV-2-encoded Vpx with the transcription factor RelA (also called p65). For the latter interaction, we documented a functional relevance in antagonizing NF-κB-driven gene expression. The mutation of the DDB1 binding interface of Vpx significantly impaired NF-κB inhibition, indicating that Vpx counteracts NF-κB signaling by a DDB1- and CRL-dependent mechanism. In summary, our findings improve the understanding of how viral pathogens hijack cellular DDB1 and CRLs to ensure efficient replication despite the expression of host restriction factors.
Keywords: DNA damage-binding protein (DDB1); NF-κB; cytomegalovirus; hepatitis B virus (HBV); human immunodeficiency virus (HIV); interaction partner; interferon; mass spectrometry (MS).
Figures




References
-
- Trilling M, Bellora N, Rutkowski AJ, De Graaf M, Dickinson P, Robertson K, et al. . Deciphering the modulation of gene expression by type I and II interferons combining 4sU-tagging, translational arrest and in silico promoter analysis. Nucleic Acids Res. (2013) 41:8107–25. 10.1093/nar/gkt589 - DOI - PMC - PubMed
-
- Megger DA, Philipp J, Le-Trilling VTK, Sitek B, Trilling M. Deciphering of the human interferon-regulated proteome by mass spectrometry-based quantitative analysis reveals extent and dynamics of protein induction and repression. Front Immunol. (2017) 8:1139. 10.3389/fimmu.2017.01139 - DOI - PMC - PubMed
-
- Rock KL, Gramm C, Rothstein L, Clark K, Stein R, Dick L, et al. . Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell (1994) 78:761–71. - PubMed
-
- Hershko A, Ciechanover A. The ubiquitin system. Ann Rev Biochem. (1998) 67:425–79. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

