Mesenteric CD103+DCs Initiate Switched Coxsackievirus B3 VP1-Specific IgA Response to Intranasal Chitosan-DNA Vaccine Through Secreting BAFF/IL-6 and Promoting Th17/Tfh Differentiation
- PMID: 30619341
- PMCID: PMC6305319
- DOI: 10.3389/fimmu.2018.02986
Mesenteric CD103+DCs Initiate Switched Coxsackievirus B3 VP1-Specific IgA Response to Intranasal Chitosan-DNA Vaccine Through Secreting BAFF/IL-6 and Promoting Th17/Tfh Differentiation
Abstract
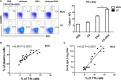
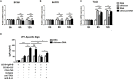
Intranasal chitosan-formulated DNA vaccination promotes IgA secretion in the intestine. However, the mechanism whereby chitosan-DNA skews IgA class switch recombination (CSR) of B cells in the Gut-associated lymph tissue (GALT) is not fully resolved. In this study, we investigated the effects of nasally administered chitosan-DNA (pcDNA3.1-VP1 plasmid encoding VP1 capsid protein of Coxsackievirus B3) on IgA production, DC activation and Tfh/Th17 response in the intestine. Compared to DNA immunization, intranasal chitosan-DNA vaccination induced antigen-specific IgA production in feces, a pronounced switching of antigen-specific IgA+ plasmablast B cells in the mesenteric lymph nodes (MLNs) and an enhanced expression of post-recombination Iα-CH transcripts/IgA germline transcript (αGT) as well as activation-induced cytidine deaminase (AID) in MLN B cells. MLN Tfh frequency was markedly enhanced by chitosan-DNA, and was associated with VP1-specific IgA titer. 24 h after immunization, intranasal chitosan-DNA induced a recruitment of CD103+DCs into the MLN that paralleled a selective loss of CD103+DCs in the lamina propria (LP). In vivo activated MLN-derived CD103+DCs produced high levels of IL-6 and BAFF in response to chitosan-DNA, which up-regulated transmembrane activator and CAML interactor (TACI) expression on MLN B cells. Upon co-culture with IgM+B in the presence of chitosan-DNA, MLN CD103+DCs induced IgA production in a T-dependent manner; and this IgA-promoting effect of CD103+DC was blocked by targeting TACI and, to a lower extent, by blocking IL-6. MLN CD103+DCs displayed an enhanced capacity to induce an enhanced CD4+Th17 response in vivo and in vitro, and IL-17A deficient mice had a pronounced reduction of specific intestinal IgA following immunization. Taken together, mesenteric CD103+DCs are indispensable for the adjuvant activity of chitosan in enhancing DNA vaccine-specific IgA switching in gut through activating BAFF-TACI and IL-6-IL-6R signaling, and through inducing Th17/Tfh differentiation in the MLN.
Keywords: CD103+DC; IgA; TACI; chitosan; class switch recombination (CSR).
Figures


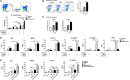

Similar articles
-
IRF4 transcription-factor-dependent CD103(+)CD11b(+) dendritic cells drive mucosal T helper 17 cell differentiation.Immunity. 2013 May 23;38(5):958-69. doi: 10.1016/j.immuni.2013.03.009. Epub 2013 May 9. Immunity. 2013. PMID: 23664832
-
Systemic flagellin immunization stimulates mucosal CD103+ dendritic cells and drives Foxp3+ regulatory T cell and IgA responses in the mesenteric lymph node.J Immunol. 2012 Dec 15;189(12):5745-54. doi: 10.4049/jimmunol.1202283. Epub 2012 Nov 14. J Immunol. 2012. PMID: 23152564
-
Incorporation of a bi-functional protein FimH enhances the immunoprotection of chitosan-pVP1 vaccine against coxsackievirus B3-induced myocarditis.Antiviral Res. 2017 Apr;140:121-132. doi: 10.1016/j.antiviral.2017.01.020. Epub 2017 Jan 28. Antiviral Res. 2017. PMID: 28137624
-
Re-thinking the functions of IgA(+) plasma cells.Gut Microbes. 2014;5(5):652-62. doi: 10.4161/19490976.2014.969977. Gut Microbes. 2014. PMID: 25483334 Free PMC article. Review.
-
Mucosal immunity: an overview and studies of enteric and respiratory coronavirus infections in a swine model of enteric disease.Vet Immunol Immunopathol. 1996 Nov;54(1-4):163-9. doi: 10.1016/s0165-2427(96)05702-9. Vet Immunol Immunopathol. 1996. PMID: 8988861 Free PMC article. Review.
Cited by
-
Resolving the Paradox of Colon Cancer Through the Integration of Genetics, Immunology, and the Microbiota.Front Immunol. 2020 Dec 14;11:600886. doi: 10.3389/fimmu.2020.600886. eCollection 2020. Front Immunol. 2020. PMID: 33381121 Free PMC article. Review.
-
Chitosan-Based Nanomaterial as Immune Adjuvant and Delivery Carrier for Vaccines.Vaccines (Basel). 2022 Nov 11;10(11):1906. doi: 10.3390/vaccines10111906. Vaccines (Basel). 2022. PMID: 36423002 Free PMC article. Review.
-
IL-22 produced by Th22 cells aggravates atherosclerosis development in ApoE-/- mice by enhancing DC-induced Th17 cell proliferation.J Cell Mol Med. 2020 Mar;24(5):3064-3078. doi: 10.1111/jcmm.14967. Epub 2020 Feb 5. J Cell Mol Med. 2020. PMID: 32022386 Free PMC article.
-
Progress in the Development of Chitosan-Based Biomaterials for Tissue Engineering and Regenerative Medicine.Biomolecules. 2019 Sep 10;9(9):470. doi: 10.3390/biom9090470. Biomolecules. 2019. PMID: 31509976 Free PMC article. Review.
-
Heterologous booster vaccination enhances antibody responses to SARS-CoV-2 by improving Tfh function and increasing B-cell clonotype SHM frequency.Front Immunol. 2024 Jun 21;15:1406138. doi: 10.3389/fimmu.2024.1406138. eCollection 2024. Front Immunol. 2024. PMID: 38975334 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

