Glycosyltransferase Family 61 in Liliopsida (Monocot): The Story of a Gene Family Expansion
- PMID: 30619412
- PMCID: PMC6297846
- DOI: 10.3389/fpls.2018.01843
Glycosyltransferase Family 61 in Liliopsida (Monocot): The Story of a Gene Family Expansion
Abstract
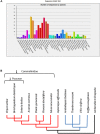
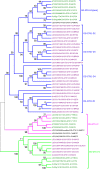
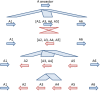
Plant cell walls play a fundamental role in several plant traits and also influence crop use as livestock nutrition or biofuel production. The Glycosyltransferase family 61 (GT61) is involved in the synthesis of cell wall xylans. In grasses (Poaceae), a copy number expansion was reported for the GT61 family, and raised the question of the evolutionary history of this gene family in a broader taxonomic context. A phylogenetic study was performed on GT61 members from 13 species representing the major angiosperm clades, in order to classify the genes, reconstruct the evolutionary history of this gene family and study its expansion in monocots. Four orthogroups (OG) were identified in angiosperms with two of them displaying a copy number expansion in monocots. These copy number expansions resulted from both tandem and segmental duplications during the genome evolution of monocot lineages. Positive selection footprints were detected on the ancestral branch leading to one of the orthogroups suggesting that the gene number expansion was accompanied by functional diversification, at least partially. We propose an OG-based classification framework for the GT61 genes at different taxonomic levels of the angiosperm useful for any further functional or translational biology study.
Keywords: Liliopsida; gene family expansion; glycosyltransferase family 61; orthologous genes; phylogeny; positive selection footprints.
Figures



References
LinkOut - more resources
Full Text Sources
Miscellaneous

