Predicting Gleason Score of Prostate Cancer Patients Using Radiomic Analysis
- PMID: 30619764
- PMCID: PMC6305278
- DOI: 10.3389/fonc.2018.00630
Predicting Gleason Score of Prostate Cancer Patients Using Radiomic Analysis
Abstract
Purpose: Use of quantitative imaging features and encoding the intra-tumoral heterogeneity from multi-parametric magnetic resonance imaging (mpMRI) for the prediction of Gleason score is gaining attention as a non-invasive biomarker for prostate cancer (PCa). This study tested the hypothesis that radiomic features, extracted from mpMRI, could predict the Gleason score pattern of patients with PCa. Methods: This analysis included T2-weighted (T2-WI) and apparent diffusion coefficient (ADC, computed from diffusion-weighted imaging) scans of 99 PCa patients from The Cancer Imaging Archive (TCIA). A total of 41 radiomic features were calculated from a local tumor sub-volume (i.e., regions of interest) that is determined by a centroid coordinate of PCa volume, grouped based on their Gleason score patterns. Kruskal-Wallis and Spearman's rank correlation tests were used to identify features related to Gleason score groups. Random forest (RF) classifier model was used to predict Gleason score groups and identify the most important signature among the 41 radiomic features. Results: Gleason score groups could be discriminated based on zone size percentage, large zone size emphasis and zone size non-uniformity values (p < 0.05). These features also showed a significant correlation between radiomic features and Gleason score groups with a correlation value of -0.35, 0.32, 0.42 for the large zone size emphasis, zone size non-uniformity and zone size percentage, respectively (corrected p < 0.05). RF classifier model achieved an average of the area under the curves of the receiver operating characteristic (ROC) of 83.40, 72.71, and 77.35% to predict Gleason score groups (G1) = 6; 6 < (G2) < (3 + 4) and (G3) ≥ 4 + 3, respectively. Conclusion: Our results suggest that the radiomic features can be used as a non-invasive biomarker to predict the Gleason score of the PCa patients.
Keywords: biomarkers; classification; gleason score; prostate cancer; radiomics.
Figures

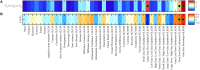

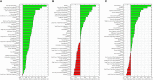

References
-
- Bard RL, Fütterer JJ, Sperling D. Image Guided Prostate Cancer Treatments. Berlin; Heidelberg: Springer; (2014).
LinkOut - more resources
Full Text Sources

