Optimized ChIP-seq method facilitates transcription factor profiling in human tumors
- PMID: 30620009
- PMCID: PMC6311467
- DOI: 10.26508/lsa.201800115
Optimized ChIP-seq method facilitates transcription factor profiling in human tumors
Abstract
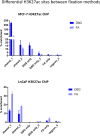
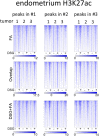
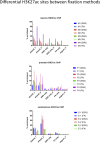
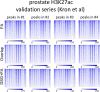
Chromatin immunoprecipitation (ChIP)-seq analyses of transcription factors in clinical specimens are challenging due to the technical limitations and low quantities of starting material, often resulting in low enrichments and poor signal-to-noise ratio. Here, we present an optimized protocol for transcription factor ChIP-seq analyses in human tissue, yielding an ∼100% success rate for all transcription factors analyzed. As proof of concept and to illustrate general applicability of the approach, human tissue from the breast, prostate, and endometrial cancers were analyzed. In addition to standard formaldehyde fixation, disuccinimidyl glutarate was included in the procedure, greatly increasing data quality. To illustrate the sensitivity of the optimized protocol, we provide high-quality ChIP-seq data for three independent factors (AR, FOXA1, and H3K27ac) from a single core needle prostate cancer biopsy specimen. In summary, double-cross-linking strongly improved transcription factor ChIP-seq quality on human tumor samples, further facilitating and enhancing translational research on limited amounts of tissue.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures




















References
-
- Droog M, Nevedomskaya E, Dackus GM, Fles R, Kim Y, Hollema H, Mourits MJ, Nederlof PM, van Boven HH, Linn SC, et al. (2017) Estrogen receptor alpha wields treatment-specific enhancers between morphologically similar endometrial tumors. Proc Natl Acad Sci U S A 114: E1316–E1325. 10.1073/pnas.1615233114 - DOI - PMC - PubMed
-
- Engelen E, Brandsma JH, Moen MJ, Signorile L, Dekkers DH, Demmers J, Kockx CE, Ozgur Z, van IWF, van den Berg DL, et al. (2015) Proteins that bind regulatory regions identified by histone modification chromatin immunoprecipitations and mass spectrometry. Nat Commun 6: 7155 10.1038/ncomms8155 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
