Reciprocal control of motility and biofilm formation by the PdhS2 two-component sensor kinase of Agrobacterium tumefaciens
- PMID: 30620265
- PMCID: PMC7003649
- DOI: 10.1099/mic.0.000758
Reciprocal control of motility and biofilm formation by the PdhS2 two-component sensor kinase of Agrobacterium tumefaciens
Abstract
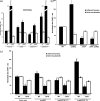
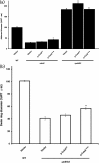
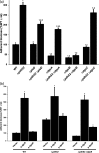
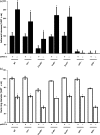
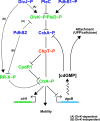
A core regulatory pathway that directs developmental transitions and cellular asymmetries in Agrobacterium tumefaciens involves two overlapping, integrated phosphorelays. One of these phosphorelays putatively includes four histidine sensor kinase homologues, DivJ, PleC, PdhS1 and PdhS2, and two response regulators, DivK and PleD. In several different alphaproteobacteria, this pathway influences a conserved downstream phosphorelay that ultimately controls the phosphorylation state of the CtrA master response regulator. The PdhS2 sensor kinase reciprocally regulates biofilm formation and swimming motility. In the current study, the mechanisms by which the A. tumefaciens sensor kinase PdhS2 directs this regulation are delineated. PdhS2 lacking a key residue implicated in phosphatase activity is markedly deficient in proper control of attachment and motility phenotypes, whereas a kinase-deficient PdhS2 mutant is only modestly affected. A genetic interaction between DivK and PdhS2 is revealed, unmasking one of several connections between PdhS2-dependent phenotypes and transcriptional control by CtrA. Epistasis experiments suggest that PdhS2 may function independently of the CckA sensor kinase, the cognate sensor kinase for CtrA, which is inhibited by DivK. Global expression analysis of the pdhS2 mutant reveals a restricted regulon, most likely functioning through CtrA to separately control motility and regulate the levels of the intracellular signal cyclic diguanylate monophosphate (cdGMP), thereby affecting the production of adhesive polysaccharides and attachment. We hypothesize that in A. tumefaciens the CtrA regulatory circuit has expanded to include additional inputs through the addition of PdhS-type sensor kinases, likely fine-tuning the response of this organism to the soil microenvironment.
Keywords: Agrobacterium tumefaciens; biofilm; development; motility; phosphorelay; sensor kinase.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures


Similar articles
-
Coordination of division and development influences complex multicellular behavior in Agrobacterium tumefaciens.PLoS One. 2013;8(2):e56682. doi: 10.1371/journal.pone.0056682. Epub 2013 Feb 20. PLoS One. 2013. PMID: 23437210 Free PMC article.
-
The branched CcsA/CckA-ChpT-CtrA phosphorelay of Sphingomonas melonis controls motility and biofilm formation.Mol Microbiol. 2015 Jul;97(1):47-63. doi: 10.1111/mmi.13011. Epub 2015 Apr 28. Mol Microbiol. 2015. PMID: 25825287
-
The CckA-ChpT-CtrA Phosphorelay Controlling Rhodobacter capsulatus Gene Transfer Agent Production Is Bidirectional and Regulated by Cyclic di-GMP.J Bacteriol. 2021 Feb 8;203(5):e00525-20. doi: 10.1128/JB.00525-20. Print 2021 Feb 8. J Bacteriol. 2021. PMID: 33288624 Free PMC article.
-
The ChvG-ChvI Regulatory Network: A Conserved Global Regulatory Circuit Among the Alphaproteobacteria with Pervasive Impacts on Host Interactions and Diverse Cellular Processes.Annu Rev Microbiol. 2023 Sep 15;77:131-148. doi: 10.1146/annurev-micro-120822-102714. Epub 2023 Apr 11. Annu Rev Microbiol. 2023. PMID: 37040790 Review.
-
Mechanisms and regulation of polar surface attachment in Agrobacterium tumefaciens.Curr Opin Microbiol. 2009 Dec;12(6):708-14. doi: 10.1016/j.mib.2009.09.014. Epub 2009 Oct 29. Curr Opin Microbiol. 2009. PMID: 19879182 Free PMC article. Review.
Cited by
-
Loss of the Rhodobacter capsulatus Serine Acetyl Transferase Gene, cysE1, Impairs Gene Transfer by Gene Transfer Agents and Biofilm Phenotypes.Appl Environ Microbiol. 2022 Oct 11;88(19):e0094422. doi: 10.1128/aem.00944-22. Epub 2022 Sep 13. Appl Environ Microbiol. 2022. PMID: 36098534 Free PMC article.
-
Regulation of Bacterial Cell Cycle Progression by Redundant Phosphatases.J Bacteriol. 2020 Aug 10;202(17):e00345-20. doi: 10.1128/JB.00345-20. Print 2020 Aug 10. J Bacteriol. 2020. PMID: 32571969 Free PMC article.
-
Transcriptome architecture of the three main lineages of agrobacteria.mSystems. 2023 Aug 31;8(4):e0033323. doi: 10.1128/msystems.00333-23. Epub 2023 Jul 21. mSystems. 2023. PMID: 37477440 Free PMC article.
-
LD-transpeptidation is crucial for fitness and polar growth in Agrobacterium tumefaciens.PLoS Genet. 2024 Oct 21;20(10):e1011449. doi: 10.1371/journal.pgen.1011449. eCollection 2024 Oct. PLoS Genet. 2024. PMID: 39432536 Free PMC article.
-
Genome-centric metagenomics reveals insights into the evolution and metabolism of a new free-living group in Rhizobiales.BMC Microbiol. 2021 Oct 28;21(1):294. doi: 10.1186/s12866-021-02354-4. BMC Microbiol. 2021. PMID: 34711170 Free PMC article.
References
-
- Dworkin M. Developmental Biology of the Bacteria. Reading, Mass: Benjamin/Cummings Pub. Co; 1985.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

