Immune genes are hotspots of shared positive selection across birds and mammals
- PMID: 30620335
- PMCID: PMC6338464
- DOI: 10.7554/eLife.41815
Immune genes are hotspots of shared positive selection across birds and mammals
Abstract
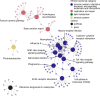
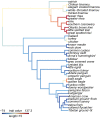
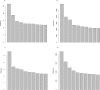
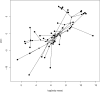
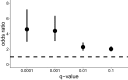
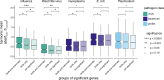
Consistent patterns of positive selection in functionally similar genes can suggest a common selective pressure across a group of species. We use alignments of orthologous protein-coding genes from 39 species of birds to estimate parameters related to positive selection for 11,000 genes conserved across birds. We show that functional pathways related to the immune system, recombination, lipid metabolism, and phototransduction are enriched for positively selected genes. By comparing our results with mammalian data, we find a significant enrichment for positively selected genes shared between taxa, and that these shared selected genes are enriched for viral immune pathways. Using pathogen-challenge transcriptome data, we show that genes up-regulated in response to pathogens are also enriched for positively selected genes. Together, our results suggest that pathogens, particularly viruses, consistently target the same genes across divergent clades, and that these genes are hotspots of host-pathogen conflict over deep evolutionary time.
Keywords: birds; chicken; comparative genomics; comparative transcriptomics; evolutionary biology; genetics; genomics; host-pathogen co-evolution; mammals; viruses.
© 2019, Shultz and Sackton.
Conflict of interest statement
AS, TS No competing interests declared
Figures









References
-
- Andersson MN, Wang H-L, Nord A, Salmón P, Isaksson C. Composition of physiologically important fatty acids in great tits differs between urban and rural populations on a seasonal basis. Frontiers in Ecology and Evolution. 2015;3:522. doi: 10.3389/fevo.2015.00093. - DOI
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

