FKBP25 participates in DNA double-strand break repair
- PMID: 30620620
- PMCID: PMC7457334
- DOI: 10.1139/bcb-2018-0328
FKBP25 participates in DNA double-strand break repair
Abstract
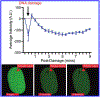
FK506-binding proteins (FKBPs) alter the conformation of proteins via cis-trans isomerization of prolyl-peptide bonds. While this activity can be demonstrated in vitro, the intractability of detecting prolyl isomerization events in cells has limited our understanding of the biological processes regulated by FKBPs. Here we report that FKBP25 is an active participant in the repair of DNA double-strand breaks (DSBs). FKBP25 influences DSB repair pathway choice by promoting homologous recombination (HR) and suppressing single-strand annealing (SSA). Consistent with this observation, cells depleted of FKBP25 form fewer Rad51 repair foci in response to etoposide and ionizing radiation, and they are reliant on the SSA repair factor Rad52 for viability. We find that FKBP25's catalytic activity is required for promoting DNA repair, which is the first description of a biological function for this enzyme activity. Consistent with the importance of the FKBP catalytic site in HR, rapamycin treatment also impairs homologous recombination, and this effect is at least in part independent of mTor. Taken together these results identify FKBP25 as a component of the DNA DSB repair pathway.
Les protéines de liaison du FK506 (FKBP) modifient la conformation des protéines au moyen de l’isomérisation cis-trans des liens peptidiques de la proline. Alors que cette activité peut être démontrée in vitro, la grande difficulté à détecter les manifestations de l’isomérisation de la proline dans les cellules a limité notre compréhension des processus biologiques régulés par les FKBP. Les auteurs rapportent ici que la FKBP25 est un participant actif dans la réparation des bris double brin (BDB) d’ADN. La FKBP25 influence le choix de la voie de réparation des BDB en favorisant la recombinaison homologue (RH) et en supprimant la renaturation simple brin. En accord avec cette observation, les cellules dépourvues de FKBP25 forment moins de foyers de réparation comportant Rad51 en réponse à l’étoposide et à la radiation ionisante, et elles dépendent du facteur de réparation-recombinaison Rad52 pour assurer leur viabilité. Ils ont trouvé que l’activité catalytique de FKBP25 est requise pour favoriser la réparation d’ADN, ce qui constitue la première description de la fonction biologique de l’activité de cette enzyme. En accord avec l’importance du site catalytique de FKBP dans la RH, le traitement à la rapamycine diminue aussi la recombinaison homologue et cet effet est au moins partiellement indépendant de mTor. Dans leur ensemble, ces résultats identifient la FKBP25 comme élément de la voie de réparation des BDB de l’ADN. [Traduit par la Rédaction]
Keywords: DNA repair; FKBP; homologous recombination; olaparib; prolyl isomerase; prolyl isomérase; recombinaison homologue; réparation d’ADN.
Figures

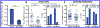



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

