Longer lifespan in the Rpd3 and Loco signaling results from the reduced catabolism in young age with noncoding RNA
- PMID: 30620723
- PMCID: PMC6339784
- DOI: 10.18632/aging.101744
Longer lifespan in the Rpd3 and Loco signaling results from the reduced catabolism in young age with noncoding RNA
Abstract
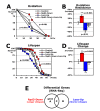
Downregulation of Rpd3 (histone deacetylase) or Loco (regulator of G-protein signaling protein) extends Drosophila lifespan with higher stress resistance. We found rpd3-downregulated long-lived flies genetically interact with loco-upregulated short-lived flies in stress resistance and lifespan. Gene expression profiles between those flies revealed that they regulate common target genes in metabolic enzymes and signaling pathways, showing an opposite expression pattern in their contrasting lifespans. Functional analyses of more significantly changed genes indicated that the activities of catabolic enzymes and uptake/storage proteins are reduced in long-lived flies with Rpd3 downregulation. This reduced catabolism exhibited from a young age is considered to be necessary for the resultant longer lifespan of the Rpd3- and Loco-downregulated old flies, which mimics the dietary restriction (DR) effect that extends lifespan in the several species. Inversely, those catabolic activities that break down carbohydrates, lipids, and peptides were high in the short lifespan of Loco-upregulated flies. Long noncoding gene, dntRL (CR45923), was also found as a putative target modulated by Rpd3 and Loco for the longevity. Interestingly, this dntRL could affect stress resistance and lifespan, suggesting that the dntRL lncRNA may be involved in the metabolic mechanism of Rpd3 and Loco signaling.
Keywords: Rpd3 and Loco signaling; lifespan; noncoding RNA; reduced catabolism; stress resistance.
Conflict of interest statement
Figures

Similar articles
-
Heart-specific Rpd3 downregulation enhances cardiac function and longevity.Aging (Albany NY). 2015 Sep;7(9):648-63. doi: 10.18632/aging.100806. Aging (Albany NY). 2015. PMID: 26399365 Free PMC article.
-
RPD3 histone deacetylase and nutrition have distinct but interacting effects on Drosophila longevity.Aging (Albany NY). 2015 Dec;7(12):1112-29. doi: 10.18632/aging.100856. Aging (Albany NY). 2015. PMID: 26647291 Free PMC article.
-
Rpd3 interacts with insulin signaling in Drosophila longevity extension.Aging (Albany NY). 2016 Nov 14;8(11):3028-3044. doi: 10.18632/aging.101110. Aging (Albany NY). 2016. PMID: 27852975 Free PMC article.
-
The effects of Rpd3 on fly metabolism, health, and longevity.Exp Gerontol. 2016 Dec 15;86:124-128. doi: 10.1016/j.exger.2016.02.015. Epub 2016 Feb 27. Exp Gerontol. 2016. PMID: 26927903 Free PMC article. Review.
-
Nutrient control of Drosophila longevity.Trends Endocrinol Metab. 2014 Oct;25(10):509-17. doi: 10.1016/j.tem.2014.02.006. Epub 2014 Mar 28. Trends Endocrinol Metab. 2014. PMID: 24685228 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

