Single-nucleotide human disease mutation inactivates a blood-regenerative GATA2 enhancer
- PMID: 30620726
- PMCID: PMC6391105
- DOI: 10.1172/JCI122694
Single-nucleotide human disease mutation inactivates a blood-regenerative GATA2 enhancer
Abstract
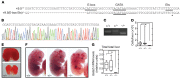
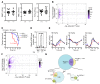
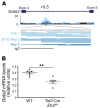
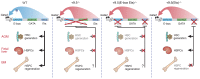
The development and function of stem and progenitor cells that produce blood cells are vital in physiology. GATA-binding protein 2 (GATA2) mutations cause GATA-2 deficiency syndrome involving immunodeficiency, myelodysplastic syndrome, and acute myeloid leukemia. GATA-2 physiological activities necessitate that it be strictly regulated, and cell type-specific enhancers fulfill this role. The +9.5 intronic enhancer harbors multiple conserved cis-elements, and germline mutations of these cis-elements are pathogenic in humans. Since mechanisms underlying how GATA2 enhancer disease mutations impact hematopoiesis and pathology are unclear, we generated mouse models of the enhancer mutations. While a multi-motif mutant was embryonically lethal, a single-nucleotide Ets motif mutant was viable, and steady-state hematopoiesis was normal. However, the Ets motif mutation abrogated stem/progenitor cell regeneration following stress. These results reveal a new mechanism in human genetics, in which a disease predisposition mutation inactivates enhancer regenerative activity, while sparing developmental activity. Mutational sensitization to stress that instigates hematopoietic failure constitutes a paradigm for GATA-2 deficiency syndrome and other contexts of GATA-2-dependent pathogenesis.
Keywords: Bone marrow differentiation; Hematology; Hematopoietic stem cells; Mouse models; Stem cells.
Conflict of interest statement
Figures