Neurog3-Independent Methylation Is the Earliest Detectable Mark Distinguishing Pancreatic Progenitor Identity
- PMID: 30620902
- PMCID: PMC6327977
- DOI: 10.1016/j.devcel.2018.11.048
Neurog3-Independent Methylation Is the Earliest Detectable Mark Distinguishing Pancreatic Progenitor Identity
Abstract
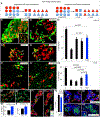
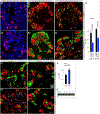
In the developing pancreas, transient Neurog3-expressing progenitors give rise to four major islet cell types: α, β, δ, and γ; when and how the Neurog3+ cells choose cell fate is unknown. Using single-cell RNA-seq, trajectory analysis, and combinatorial lineage tracing, we showed here that the Neurog3+ cells co-expressing Myt1 (i.e., Myt1+Neurog3+) were biased toward β cell fate, while those not simultaneously expressing Myt1 (Myt1-Neurog3+) favored α fate. Myt1 manipulation only marginally affected α versus β cell specification, suggesting Myt1 as a marker but not determinant for islet-cell-type specification. The Myt1+Neurog3+ cells displayed higher Dnmt1 expression and enhancer methylation at Arx, an α-fate-promoting gene. Inhibiting Dnmts in pancreatic progenitors promoted α cell specification, while Dnmt1 overexpression or Arx enhancer hypermethylation favored β cell production. Moreover, the pancreatic progenitors contained distinct Arx enhancer methylation states without transcriptionally definable sub-populations, a phenotype independent of Neurog3 activity. These data suggest that Neurog3-independent methylation on fate-determining gene enhancers specifies distinct endocrine-cell programs.
Keywords: Arx; DMR; DNA methylation; DNMT; HMR; Myt1; azacytidine; combinatorial lineage tracing; diabetes; epigenetics; glucagon; insulin; lineage priming; p-Creode; pseudotime; single-cell RNA-seq; specification; stochastic gene expression; trajectory; transcriptional noise; α cell; β cell.
Published by Elsevier Inc.
Conflict of interest statement
Declarations of Interests:
The authors declare no competing interests.
Figures





References
-
- Bastidas-Ponce A, Scheibner K, Lickert H, and Bakhti M (2017). Cellular and molecular mechanisms coordinating pancreas development. Development 144, 2873–2888. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32 HD007502/HD/NICHD NIH HHS/United States
- P50 CA095103/CA/NCI NIH HHS/United States
- U54 MD007586/MD/NIMHD NIH HHS/United States
- P30 DK058404/DK/NIDDK NIH HHS/United States
- R01 DK106228/DK/NIDDK NIH HHS/United States
- U2C DK059637/DK/NIDDK NIH HHS/United States
- R01 DK065949/DK/NIDDK NIH HHS/United States
- P30 EY008126/EY/NEI NIH HHS/United States
- P30 DK020593/DK/NIDDK NIH HHS/United States
- P30 CA068485/CA/NCI NIH HHS/United States
- F31 GM120940/GM/NIGMS NIH HHS/United States
- R01 DK103831/DK/NIDDK NIH HHS/United States
- U24 DK059637/DK/NIDDK NIH HHS/United States
- R01 DK125696/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

