Cyclin C: The Story of a Non-Cycling Cyclin
- PMID: 30621145
- PMCID: PMC6466611
- DOI: 10.3390/biology8010003
Cyclin C: The Story of a Non-Cycling Cyclin
Abstract
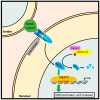
The class I cyclin family is a well-studied group of structurally conserved proteins that interact with their associated cyclin-dependent kinases (Cdks) to regulate different stages of cell cycle progression depending on their oscillating expression levels. However, the role of class II cyclins, which primarily act as transcription factors and whose expression remains constant throughout the cell cycle, is less well understood. As a classic example of a transcriptional cyclin, cyclin C forms a regulatory sub-complex with its partner kinase Cdk8 and two accessory subunits Med12 and Med13 called the Cdk8-dependent kinase module (CKM). The CKM reversibly associates with the multi-subunit transcriptional coactivator complex, the Mediator, to modulate RNA polymerase II-dependent transcription. Apart from its transcriptional regulatory function, recent research has revealed a novel signaling role for cyclin C at the mitochondria. Upon oxidative stress, cyclin C leaves the nucleus and directly activates the guanosine 5'-triphosphatase (GTPase) Drp1, or Dnm1 in yeast, to induce mitochondrial fragmentation. Importantly, cyclin C-induced mitochondrial fission was found to increase sensitivity of both mammalian and yeast cells to apoptosis. Here, we review and discuss the biology of cyclin C, focusing mainly on its transcriptional and non-transcriptional roles in tumor promotion or suppression.
Keywords: Cdk8-dependent kinase module; Mediator; cancer; cyclin family; stress signaling; transcriptional cyclins; tumor suppressor; yeast.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

