Vibrational Approach to the Dynamics and Structure of Protein Amyloids
- PMID: 30621325
- PMCID: PMC6337179
- DOI: 10.3390/molecules24010186
Vibrational Approach to the Dynamics and Structure of Protein Amyloids
Abstract
Amyloid diseases, including neurodegenerative diseases such as Alzheimer's and Parkinson's, are linked to a poorly understood progression of protein misfolding and aggregation events that culminate in tissue-selective deposition and human pathology. Elucidation of the mechanistic details of protein aggregation and the structural features of the aggregates is critical for a comprehensive understanding of the mechanisms of protein oligomerization and fibrillization. Vibrational spectroscopies, such as Fourier transform infrared (FTIR) and Raman, are powerful tools that are sensitive to the secondary structure of proteins and have been widely used to investigate protein misfolding and aggregation. We address the application of the vibrational approaches in recent studies of conformational dynamics and structural characteristics of protein oligomers and amyloid fibrils. In particular, introduction of isotope labelled carbonyl into a peptide backbone, and incorporation of the extrinsic unnatural amino acids with vibrational moieties on the side chain, have greatly expanded the ability of vibrational spectroscopy to obtain site-specific structural and dynamic information. The applications of these methods in recent studies of protein aggregation are also reviewed.
Keywords: Raman; amyloid; infrared; isotopic labelling; oligomers; protein aggregation; site-specific probe; vibrational spectroscopy.
Conflict of interest statement
The authors state that they have no conflicts of interest.
Figures


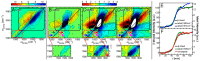
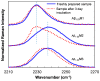
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

