Characterization of the responses of the caspase 2, 3, 6 and 8 genes to immune challenges and extracellular ATP stimulation in the Japanese flounder (Paralichthys olivaceus)
- PMID: 30621683
- PMCID: PMC6325855
- DOI: 10.1186/s12917-018-1763-y
Characterization of the responses of the caspase 2, 3, 6 and 8 genes to immune challenges and extracellular ATP stimulation in the Japanese flounder (Paralichthys olivaceus)
Abstract
Background: Caspases are a family of conserved intracellular cysteine-dependent aspartate-specific cysteine proteases that play important roles in regulating cell death and inflammation. Our previous study revealed the importance of the inflammatory caspase 1 gene in extracellular ATP-mediated immune signaling in Japanese flounder, Paralichthys olivaceus. To explore the potential roles of other caspases in P. olivaceus innate immunity, we extended our study by characterizing of the responses of four additional P. olivaceus caspase genes, termed JfCaspase 2, 3, 6 and 8, to inflammatory challenge and extracellular ATP stimulation.
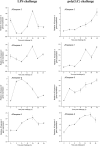
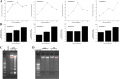
Results: Sequence analysis revealed that the domain structures of all the Japanese flounder caspase proteins are evolutionarily conserved. Quantitative real-time PCR analysis showed that the JfCaspase 2, 3, 6 and 8 genes were expressed ubiquitously but at unequal levels in all examined Japanese flounder normal tissues. In addition, the basal gene expression levels of JfCaspase 2, 3, 6 and 8 were higher than those of JfCaspase 1 in both Japanese flounder head kidney macrophages (HKMs) and peripheral blood leukocytes (PBLs). Furthermore, immune challenge experiments showed that the inflammatory stimuli LPS and poly(I:C) significantly modulated the expression of the JfCaspase 2, 3, 6 and 8 genes in Japanese flounder immune cells. Finally, DNA fragmentation, associated with increased extracellular ATP-induced JfCaspase 2, 3, 6 and 8 gene expression and enzymatic activity, was inhibited by the caspase inhibitor Z-VAD-FMK in the HKMs.
Conclusion: Our findings demonstrate broad participation of multiple caspase genes in response to inflammatory stimulation in Japanese flounder immune cells and provide new evidence for the involvement of caspase(s) in extracellular ATP-induced apoptosis in fish.
Keywords: Apoptosis; Caspases; Extracellular ATP; Immune response; Paralichthys olivaceus; Teleost.
Conflict of interest statement
Ethics approval and consent to participate
All experiments were conducted in accordance with the NIH guidelines for the care and use of experimental animals and these studies were approved by the animal care and use committee of the College of Life Sciences, Tianjin Normal University (#2018–01).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures




Similar articles
-
Identification and characterization of a novel NOD-like receptor family CARD domain containing 3 gene in response to extracellular ATP stimulation and its role in regulating LPS-induced innate immune response in Japanese flounder (Paralichthys olivaceus) head kidney macrophages.Fish Shellfish Immunol. 2016 Mar;50:79-90. doi: 10.1016/j.fsi.2016.01.029. Epub 2016 Jan 25. Fish Shellfish Immunol. 2016. PMID: 26820104
-
Characterization of Japanese flounder (Paralichthys olivaceus) Caspase1 involved in extracellular ATP-mediated immune signaling in fish.Fish Shellfish Immunol. 2017 Aug;67:536-545. doi: 10.1016/j.fsi.2017.06.043. Epub 2017 Jun 16. Fish Shellfish Immunol. 2017. PMID: 28630015
-
Expression and functional characterization of vitronectin gene from Japanese flounder (Paralichthys olivaceus).Fish Shellfish Immunol. 2017 Jun;65:9-16. doi: 10.1016/j.fsi.2017.03.055. Epub 2017 Apr 1. Fish Shellfish Immunol. 2017. PMID: 28377270
-
Caspase-8 as a regulator of tumor cell motility.Curr Mol Med. 2014 Feb;14(2):246-54. doi: 10.2174/1566524014666140128111951. Curr Mol Med. 2014. PMID: 24467204 Free PMC article. Review.
-
Caspase-8: Arbitrating Life and Death in the Innate Immune System.Cells. 2025 Feb 7;14(4):240. doi: 10.3390/cells14040240. Cells. 2025. PMID: 39996713 Free PMC article. Review.
Cited by
-
Memantine, Simvastatin, and Epicatechin Inhibit 7-Ketocholesterol-induced Apoptosis in Retinal Pigment Epithelial Cells But Not Neurosensory Retinal Cells In Vitro.J Ophthalmic Vis Res. 2020 Oct 25;15(4):470-480. doi: 10.18502/jovr.v15i4.7781. eCollection 2020 Oct-Dec. J Ophthalmic Vis Res. 2020. PMID: 33133437 Free PMC article.
-
A novel framework integrating AI model and enzymological experiments promotes identification of SARS-CoV-2 3CL protease inhibitors and activity-based probe.Brief Bioinform. 2021 Nov 5;22(6):bbab301. doi: 10.1093/bib/bbab301. Brief Bioinform. 2021. PMID: 34368837 Free PMC article.
-
Animal Models for the Investigation of P2X7 Receptors.Int J Mol Sci. 2023 May 4;24(9):8225. doi: 10.3390/ijms24098225. Int J Mol Sci. 2023. PMID: 37175933 Free PMC article. Review.
-
Toxicity Evaluation, Oxidative, and Immune Responses of Mercury on Nile Tilapia: Modulatory Role of Dietary Nannochloropsis oculata.Biol Trace Elem Res. 2024 Apr;202(4):1752-1766. doi: 10.1007/s12011-023-03771-4. Epub 2023 Jul 25. Biol Trace Elem Res. 2024. PMID: 37491615 Free PMC article.
-
Activated P2X receptors can up-regulate the expressions of inflammation-related genes via NF-κB pathway in spotted sea bass (Lateolabrax maculatus).Front Immunol. 2023 May 4;14:1181067. doi: 10.3389/fimmu.2023.1181067. eCollection 2023. Front Immunol. 2023. PMID: 37215129 Free PMC article.
References
-
- Lamkanfi M, Declercq W, Kalai M, Saelens X, Vandenabeele P. Alice in caspase land. A phylogenetic analysis of caspases from worm to man. Cell Death Differ. 2002;9(4):358–361. - PubMed
-
- Boatright KM, Salvesen GS. Mechanisms of caspase activation. Curr Opin Cell Biol. 2003;15(6):725–731. - PubMed
-
- López-Castejón G, Sepulcre MP, Mulero I, Pelegrín P, Meseguer J, Mulero V. Molecular and functional characterization of gilthead seabream Sparus aurata caspase-1: the first identification of an inflammatory caspase in fish. Mol Immunol. 2008;45(1):49–57. - PubMed
MeSH terms
Substances
Grants and funding
- 31572645/the National Natural Science Foundation of China
- NA/the University Yong Talent Cultivation Program provided by Tianjin Municipal Education Commission
- 52XC1503/the Science Promotion Program for Young Scholars provided by Tianjin Normal University
- 18JCZDJC33600/the Natural Science Foundation of Tianjin
LinkOut - more resources
Full Text Sources

