Systematic interrogation of human promoters
- PMID: 30622120
- PMCID: PMC6360817
- DOI: 10.1101/gr.236075.118
Systematic interrogation of human promoters
Abstract
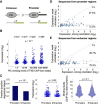
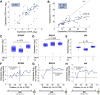
Despite much research, our understanding of the architecture and cis-regulatory elements of human promoters is still lacking. Here, we devised a high-throughput assay to quantify the activity of approximately 15,000 fully designed sequences that we integrated and expressed from a fixed location within the human genome. We used this method to investigate thousands of native promoters and preinitiation complex (PIC) binding regions followed by in-depth characterization of the sequence motifs underlying promoter activity, including core promoter elements and TF binding sites. We find that core promoters drive transcription mostly unidirectionally and that sequences originating from promoters exhibit stronger activity than those originating from enhancers. By testing multiple synthetic configurations of core promoter elements, we dissect the motifs that positively and negatively regulate transcription as well as the effect of their combinations and distances, including a 10-bp periodicity in the optimal distance between the TATA and the initiator. By comprehensively screening 133 TF binding sites, we find that in contrast to core promoters, TF binding sites maintain similar activity levels in both orientations, supporting a model by which divergent transcription is driven by two distinct unidirectional core promoters sharing bidirectional TF binding sites. Finally, we find a striking agreement between the effect of binding site multiplicity of individual TFs in our assay and their tendency to appear in homotypic clusters throughout the genome. Overall, our study systematically assays the elements that drive expression in core and proximal promoter regions and sheds light on organization principles of regulatory regions in the human genome.
© 2019 Weingarten-Gabbay et al.; Published by Cold Spring Harbor Laboratory Press.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
