Antibody Repertoire Analysis of Hepatitis C Virus Infections Identifies Immune Signatures Associated With Spontaneous Clearance
- PMID: 30622532
- PMCID: PMC6308210
- DOI: 10.3389/fimmu.2018.03004
Antibody Repertoire Analysis of Hepatitis C Virus Infections Identifies Immune Signatures Associated With Spontaneous Clearance
Abstract
Hepatitis C virus (HCV) is a major public health concern, with over 70 million people infected worldwide, who are at risk for developing life-threatening liver disease. No vaccine is available, and immunity against the virus is not well-understood. Following the acute stage, HCV usually causes chronic infections. However, ~30% of infected individuals spontaneously clear the virus. Therefore, using HCV as a model for comparing immune responses between spontaneous clearer (SC) and chronically infected (CI) individuals may empower the identification of mechanisms governing viral infection outcomes. Here, we provide the first in-depth analysis of adaptive immune receptor repertoires in individuals with current or past HCV infection. We demonstrate that SC individuals, in contrast to CI patients, develop clusters of antibodies with distinct properties. These antibodies' characteristics were used in a machine learning framework to accurately predict infection outcome. Using combinatorial antibody phage display library technology, we identified HCV-specific antibody sequences. By integrating these data with the repertoire analysis, we constructed two antibodies characterized by high neutralization breadth, which are associated with clearance. This study provides insight into the nature of effective immune response against HCV and demonstrates an innovative approach for constructing antibodies correlating with successful infection clearance. It may have clinical implications for prognosis of the future status of infection, and the design of effective immunotherapies and a vaccine for HCV.
Keywords: antibody repertoire; hepatitis C virus; immune signature; infectious disease; neutralizing antibodies.
Figures

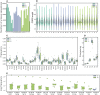
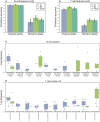
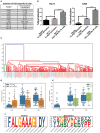
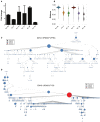
Similar articles
-
Broad Anti-Hepatitis C Virus (HCV) Antibody Responses Are Associated with Improved Clinical Disease Parameters in Chronic HCV Infection.J Virol. 2016 Apr 14;90(9):4530-4543. doi: 10.1128/JVI.02669-15. Print 2016 May. J Virol. 2016. PMID: 26912610 Free PMC article.
-
Cross-genotype AR3-specific neutralizing antibodies confer long-term protection in injecting drug users after HCV clearance.J Hepatol. 2019 Jul;71(1):14-24. doi: 10.1016/j.jhep.2019.02.013. Epub 2019 Feb 21. J Hepatol. 2019. PMID: 30797052
-
Role of the E2 Hypervariable Region (HVR1) in the Immunogenicity of a Recombinant Hepatitis C Virus Vaccine.J Virol. 2018 May 14;92(11):e02141-17. doi: 10.1128/JVI.02141-17. Print 2018 Jun 1. J Virol. 2018. PMID: 29540595 Free PMC article.
-
The hepatitis C virus glycan shield and evasion of the humoral immune response.Viruses. 2011 Oct;3(10):1909-32. doi: 10.3390/v3101909. Epub 2011 Oct 14. Viruses. 2011. PMID: 22069522 Free PMC article. Review.
-
Neutralizing antibodies and pathogenesis of hepatitis C virus infection.Viruses. 2012 Oct 9;4(10):2016-30. doi: 10.3390/v4102016. Viruses. 2012. PMID: 23202451 Free PMC article. Review.
Cited by
-
Antibody Responses in Hepatitis C Infection.Cold Spring Harb Perspect Med. 2021 Mar 1;11(3):a036962. doi: 10.1101/cshperspect.a036962. Cold Spring Harb Perspect Med. 2021. PMID: 32341067 Free PMC article. Review.
-
The Neutralizing Antibody Responses of Individuals That Spontaneously Resolve Hepatitis C Virus Infection.Viruses. 2022 Jun 25;14(7):1391. doi: 10.3390/v14071391. Viruses. 2022. PMID: 35891372 Free PMC article.
-
IGHV allele similarity clustering improves genotype inference from adaptive immune receptor repertoire sequencing data.Nucleic Acids Res. 2023 Sep 8;51(16):e86. doi: 10.1093/nar/gkad603. Nucleic Acids Res. 2023. PMID: 37548401 Free PMC article.
-
Broadly Neutralizing Antibody Characteristics in Hepatitis C Virus Infection and Implications for Vaccine Design.Vaccines (Basel). 2025 Jun 6;13(6):612. doi: 10.3390/vaccines13060612. Vaccines (Basel). 2025. PMID: 40573943 Free PMC article. Review.
-
Guidelines for reproducible analysis of adaptive immune receptor repertoire sequencing data.Brief Bioinform. 2024 Mar 27;25(3):bbae221. doi: 10.1093/bib/bbae221. Brief Bioinform. 2024. PMID: 38752856 Free PMC article.
References
-
- Trucchi C, Orsi A, Alicino C, Sticchi L, Icardi G, Ansaldi F. State of the art, unresolved issues, and future research directions in the fight against hepatitis C virus: perspectives for screening, diagnostics of resistances, and immunization. J Immunol Res. (2016) 2016:1412840. 10.1155/2016/1412840 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

