Deleterious alleles in the context of domestication, inbreeding, and selection
- PMID: 30622631
- PMCID: PMC6304688
- DOI: 10.1111/eva.12691
Deleterious alleles in the context of domestication, inbreeding, and selection
Abstract
Each individual has a certain number of harmful mutations in its genome. These mutations can lower the fitness of the individual carrying them, dependent on their dominance and selection coefficient. Effective population size, selection, and admixture are known to affect the occurrence of such mutations in a population. The relative roles of demography and selection are a key in understanding the process of adaptation. These are factors that are potentially influenced and confounded in domestic animals. Here, we hypothesize that the series of events of bottlenecks, introgression, and strong artificial selection associated with domestication increased mutational load in domestic species. Yet, mutational load is hard to quantify, so there are very few studies available revealing the relevance of evolutionary processes. The precise role of artificial selection, bottlenecks, and introgression in further increasing the load of deleterious variants in animals in breeding and conservation programmes remains unclear. In this paper, we review the effects of domestication and selection on mutational load in domestic species. Moreover, we test some hypotheses on higher mutational load due to domestication and selective sweeps using sequence data from commercial pig and chicken lines. Overall, we argue that domestication by itself is not a prerequisite for genetic erosion, indicating that fitness potential does not need to decline. Rather, mutational load in domestic species can be influenced by many factors, but consistent or strong trends are not yet clear. However, methods emerging from molecular genetics allow discrimination of hypotheses about the determinants of mutational load, such as effective population size, inbreeding, and selection, in domestic systems. These findings make us rethink the effect of our current breeding schemes on fitness of populations.
Keywords: deleterious alleles; domestication; genetic load; inbreeding; selection.
Figures
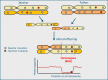



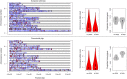
References
-
- Agrawal, A. F. , & Whitlock, M. C. (2012). Mutation load: The fitness of individuals in populations where deleterious alleles are abundant. Annual Review of Ecology, Evolution, and Systematics, 43, 115–135. 10.1146/annurev-ecolsys-110411-160257 - DOI
-
- Bosse, M. , Megens, H. J. , Madsen, O. , Frantz, L. A. F. , Paudel, Y. , Crooijmans, R. P. M. A. , & Groenen, M. A. M. (2014). Untangling the hybrid nature of modern pig genomes: A mosaic derived from biogeographically distinct and highly divergent Sus scrofa populations. Molecular Ecology, 23(16), 4089–4102. 10.1111/mec.12807 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources

