A re-evaluation of the domestication bottleneck from archaeogenomic evidence
- PMID: 30622633
- PMCID: PMC6304682
- DOI: 10.1111/eva.12680
A re-evaluation of the domestication bottleneck from archaeogenomic evidence
Abstract
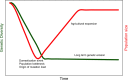
Domesticated crops show a reduced level of diversity that is commonly attributed to the "domestication bottleneck"; a drastic reduction in the population size associated with subsampling the wild progenitor species and the imposition of selection pressures associated with the domestication syndrome. A prediction of the domestication bottleneck is a sharp decline in genetic diversity early in the domestication process. Surprisingly, archaeological genomes of three major annual crops do not indicate that such a drop in diversity occurred early in the domestication process. In light of this observation, we revisit the general assumption of the domestication bottleneck concept in our current understanding of the evolutionary process of domestication.
Keywords: archaeogenomics; domestication; domestication bottleneck.
Figures

References
LinkOut - more resources
Full Text Sources

