Improved lipid production via fatty acid biosynthesis and free fatty acid recycling in engineered Synechocystis sp. PCC 6803
- PMID: 30622650
- PMCID: PMC6319012
- DOI: 10.1186/s13068-018-1349-8
Improved lipid production via fatty acid biosynthesis and free fatty acid recycling in engineered Synechocystis sp. PCC 6803
Abstract
Background: Cyanobacteria are potential sources for third generation biofuels. Their capacity for biofuel production has been widely improved using metabolically engineered strains. In this study, we employed metabolic engineering design with target genes involved in selected processes including the fatty acid synthesis (a cassette of accD, accA, accC and accB encoding acetyl-CoA carboxylase, ACC), phospholipid hydrolysis (lipA encoding lipase A), alkane synthesis (aar encoding acyl-ACP reductase, AAR), and recycling of free fatty acid (FFA) (aas encoding acyl-acyl carrier protein synthetase, AAS) in the unicellular cyanobacterium Synechocystis sp. PCC 6803.
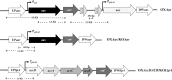
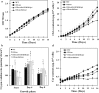
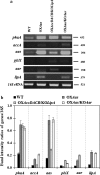
Results: To enhance lipid production, engineered strains were successfully obtained including an aas-overexpressing strain (OXAas), an aas-overexpressing strain with aar knockout (OXAas/KOAar), and an accDACB-overexpressing strain with lipA knockout (OXAccDACB/KOLipA). All engineered strains grew slightly slower than wild-type (WT), as well as with reduced levels of intracellular pigment levels of chlorophyll a and carotenoids. A higher lipid content was noted in all the engineered strains compared to WT cells, especially in OXAas, with maximal content and production rate of 34.5% w/DCW and 41.4 mg/L/day, respectively, during growth phase at day 4. The OXAccDACB/KOLipA strain, with an impediment of phospholipid hydrolysis to FFA, also showed a similarly high content of total lipid of about 32.5% w/DCW but a lower production rate of 31.5 mg/L/day due to a reduced cell growth. The knockout interruptions generated, upon a downstream flow from intermediate fatty acyl-ACP, an induced unsaturated lipid production as observed in OXAas/KOAar and OXAccDACB/KOLipA strains with 5.4% and 3.1% w/DCW, respectively.
Conclusions: Among the three metabolically engineered Synechocystis strains, the OXAas with enhanced free fatty acid recycling had the highest efficiency to increase lipid production.
Keywords: Acetyl-CoA carboxylase; Acyl-ACP reductase; Acyl–acyl carrier protein synthetase; Lipase A; Synechocystis sp. PCC 6803; Total lipid; Unsaturated lipid.
Figures



Similar articles
-
Enhanced productivity of extracellular free fatty acids by gene disruptions of acyl-ACP synthetase and S-layer protein in Synechocystis sp. PCC 6803.Biotechnol Biofuels Bioprod. 2022 Sep 24;15(1):99. doi: 10.1186/s13068-022-02197-9. Biotechnol Biofuels Bioprod. 2022. PMID: 36153604 Free PMC article.
-
Overexpression of lipA or glpD_RuBisCO in the Synechocystis sp. PCC 6803 Mutant Lacking the Aas Gene Enhances Free Fatty-Acid Secretion and Intracellular Lipid Accumulation.Int J Mol Sci. 2021 Oct 25;22(21):11468. doi: 10.3390/ijms222111468. Int J Mol Sci. 2021. PMID: 34768898 Free PMC article.
-
Synechocystis sp. PCC 6803 overexpressing genes involved in CBB cycle and free fatty acid cycling enhances the significant levels of intracellular lipids and secreted free fatty acids.Sci Rep. 2020 Mar 11;10(1):4515. doi: 10.1038/s41598-020-61100-4. Sci Rep. 2020. PMID: 32161307 Free PMC article.
-
Advancing oleaginous microorganisms to produce lipid via metabolic engineering technology.Prog Lipid Res. 2013 Oct;52(4):395-408. doi: 10.1016/j.plipres.2013.05.002. Epub 2013 May 16. Prog Lipid Res. 2013. PMID: 23685199 Review.
-
Cyanobacterial Enzymes for Bioalkane Production.Adv Exp Med Biol. 2018;1080:119-154. doi: 10.1007/978-981-13-0854-3_6. Adv Exp Med Biol. 2018. PMID: 30091094 Review.
Cited by
-
Improved lipid production and component of mycosporine-like amino acids by co-overexpression of amt1 and aroB genes in Synechocystis sp. PCC6803.Sci Rep. 2023 Nov 9;13(1):19439. doi: 10.1038/s41598-023-46290-x. Sci Rep. 2023. PMID: 37945676 Free PMC article.
-
Enhanced productivity of extracellular free fatty acids by gene disruptions of acyl-ACP synthetase and S-layer protein in Synechocystis sp. PCC 6803.Biotechnol Biofuels Bioprod. 2022 Sep 24;15(1):99. doi: 10.1186/s13068-022-02197-9. Biotechnol Biofuels Bioprod. 2022. PMID: 36153604 Free PMC article.
-
Incorporation, fate, and turnover of free fatty acids in cyanobacteria.FEMS Microbiol Rev. 2023 Mar 10;47(2):fuad015. doi: 10.1093/femsre/fuad015. FEMS Microbiol Rev. 2023. PMID: 37061785 Free PMC article. Review.
-
Untargeted Lipidomics Analysis of the Cyanobacterium Synechocystis sp. PCC 6803: Lipid Composition Variation in Response to Alternative Cultivation Setups and to Gene Deletion.Int J Mol Sci. 2020 Nov 24;21(23):8883. doi: 10.3390/ijms21238883. Int J Mol Sci. 2020. PMID: 33255174 Free PMC article.
-
Emerging Trends in Genetic Engineering of Microalgae for Commercial Applications.Mar Drugs. 2022 Apr 24;20(5):285. doi: 10.3390/md20050285. Mar Drugs. 2022. PMID: 35621936 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

