Chromosomal Microarray Analysis as a First-Tier Clinical Diagnostic Test in Patients With Developmental Delay/Intellectual Disability, Autism Spectrum Disorders, and Multiple Congenital Anomalies: A Prospective Multicenter Study in Korea
- PMID: 30623622
- PMCID: PMC6340852
- DOI: 10.3343/alm.2019.39.3.299
Chromosomal Microarray Analysis as a First-Tier Clinical Diagnostic Test in Patients With Developmental Delay/Intellectual Disability, Autism Spectrum Disorders, and Multiple Congenital Anomalies: A Prospective Multicenter Study in Korea
Abstract
Background: To validate the clinical application of chromosomal microarray analysis (CMA) as a first-tier clinical diagnostic test and to determine the impact of CMA results on patient clinical management, we conducted a multicenter prospective study in Korean patients diagnosed as having developmental delay/intellectual disability (DD/ID), autism spectrum disorders (ASD), and multiple congenital anomalies (MCA).
Methods: We performed both CMA and G-banding cytogenetics as the first-tier tests in 617 patients. To determine whether the CMA results directly influenced treatment recommendations, the referring clinicians were asked to complete a 39-item questionnaire for each patient separately after receiving the CMA results.
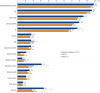
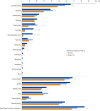
Results: A total of 122 patients (19.8%) had abnormal CMA results, with either pathogenic variants (N=65) or variants of possible significance (VPS, N=57). Thirty-five well-known diseases were detected: 16p11.2 microdeletion syndrome was the most common, followed by Prader-Willi syndrome, 15q11-q13 duplication, Down syndrome, and Duchenne muscular dystrophy. Variants of unknown significance (VUS) were discovered in 51 patients (8.3%). VUS of genes putatively associated with developmental disorders were found in five patients: IMMP2L deletion, PTCH1 duplication, and ATRNL1 deletion. CMA results influenced clinical management, such as imaging studies, specialist referral, and laboratory testing in 71.4% of patients overall, and in 86.0%, 83.3%, 75.0%, and 67.3% of patients with VPS, pathogenic variants, VUS, and benign variants, respectively.
Conclusions: Clinical application of CMA as a first-tier test improves diagnostic yields and the quality of clinical management in patients with DD/ID, ASD, and MCA.
Keywords: Autism spectrum disorders; Benign; Chromosomal microarray analysis; Clinical management; Developmental delay; Intellectual disability; Multiple congenital anomalies; Pathogenic; Variant of possible significance; Variant of unknown significance.
© The Korean Society for Laboratory Medicine.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
References
-
- Sanmann JN, Pickering DL, Golden DM, Stevens JM, Hempel TE, Althof PA, et al. Assessing the utility of confirmatory studies following identification of large-scale genomic imbalances by microarray. Genet Med. 2015;17:875–879. - PubMed
-
- Haddow JE, Palomaki GE. ACCE: a model process for evaluating data on emerging genetic tests. In: Khoury MJ, Little J, Burke W, editors. Human genome epidemiology: a scientific foundation for using genetic information to improve health and prevent disease. New York: Oxford University Press; 2004. pp. 217–233.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

