PubTerm: a web tool for organizing, annotating and curating genes, diseases, molecules and other concepts from PubMed records
- PMID: 30624653
- PMCID: PMC6323318
- DOI: 10.1093/database/bay137
PubTerm: a web tool for organizing, annotating and curating genes, diseases, molecules and other concepts from PubMed records
Abstract
Background and objective: Analysis, annotation and curation of biomedical scientific literature is a recurrent task in biomedical research, database curation and clinics. Commonly, the reading is centered on concepts such as genes, diseases or molecules. Database curators may also need to annotate published abstracts related to a specific topic. However, few free and intuitive tools exist to assist users in this context. Therefore, we developed PubTerm, a web tool to organize, categorize, curate and annotate a large number of PubMed abstracts related to biological entities such as genes, diseases, chemicals, species, sequence variants and other related information.
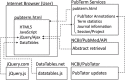
Methods: A variety of interfaces were implemented to facilitate curation and annotation, including the organization of abstracts by terms, by the co-occurrence of terms or by specific phrases. Information includes statistics on the occurrence of terms. The abstracts, terms and other related information can be annotated and categorized using user-defined categories. The session information can be saved and restored, and the data can be exported to other formats.
Results: The pipeline in PubTerm starts by specifying a PubMed query or list of PubMed identifiers. Then, the user can specify three lists of categories and specify what information will be highlighted in which colors. The user then utilizes the `term view' to organize the abstracts by gene, disease, species or other information to facilitate the annotation and categorization of terms or abstracts. Other views also facilitate the exploration of abstracts and connections between terms. We have used PubTerm to quickly and efficiently curate collections of more than 400 abstracts that mention more than 350 genes to generate revised lists of susceptibility genes for diseases. An example is provided for pulmonary arterial hypertension.
Conclusions: PubTerm saves time for literature revision by assisting with annotation organization and knowledge acquisition.
Figures




References
-
- Keepanasseril A. (2014) PubMed alternatives to search MEDLINE: an environmental scan. Indian J. Dent. Res., 25, 527. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

