Effects of altering histone posttranslational modifications on mitotic chromosome structure and mechanics
- PMID: 30625026
- PMCID: PMC6589789
- DOI: 10.1091/mbc.E18-09-0592
Effects of altering histone posttranslational modifications on mitotic chromosome structure and mechanics
Abstract
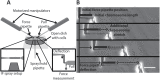
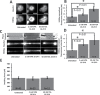
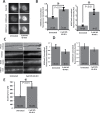
During cell division, chromatin is compacted into mitotic chromosomes to aid faithful segregation of the genome between two daughter cells. Posttranslational modifications (PTMs) of histones alter compaction of interphase chromatin, but it remains poorly understood how these modifications affect mitotic chromosome stiffness and structure. Using micropipette-based force measurements and epigenetic drugs, we probed the influence of canonical histone PTMs that dictate interphase euchromatin (acetylation) and heterochromatin (methylation) on mitotic chromosome stiffness. By measuring chromosome doubling force (the force required to double chromosome length), we find that histone methylation, but not acetylation, contributes to mitotic structure and stiffness. We discuss our findings in the context of chromatin gel modeling of the large-scale organization of mitotic chromosomes.
Figures


References
-
- Ball AR, Jr, Yokomori K. (2001). The structural maintenance of chromosomes (SMC) family of proteins in mammals. Chromosome Res , 85–96. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

