Cophylogenetic analysis suggests cospeciation between the Scorpion Mycoplasma Clade symbionts and their hosts
- PMID: 30625167
- PMCID: PMC6326461
- DOI: 10.1371/journal.pone.0209588
Cophylogenetic analysis suggests cospeciation between the Scorpion Mycoplasma Clade symbionts and their hosts
Abstract
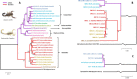
Scorpions are predator arachnids of ancient origin and worldwide distribution. Two scorpion species, Vaejovis smithi and Centruroides limpidus, were found to harbor two different Mollicutes phylotypes: a Scorpion Mycoplasma Clade (SMC) and Scorpion Group 1 (SG1). Here we investigated, using a targeted gene sequencing strategy, whether these Mollicutes were present in 23 scorpion morphospecies belonging to the Vaejovidae, Carboctonidae, Euscorpiidae, Diplocentridae, and Buthidae families. Our results revealed that SMC is found in a species-specific association with Vaejovidae and Buthidae, whereas SG1 is uniquely found in Vaejovidae. SMC and SG1 co-occur only in Vaejovis smithi where 43% of the individuals host both phylotypes. A phylogenetic analysis of Mollicutes 16S rRNA showed that SMC and SG1 constitute well-delineated phylotypes. Additionally, we found that SMC and scorpion phylogenies are significantly congruent, supporting the observation that a cospeciation process may have occurred. This study highlights the phylogenetic diversity of the scorpion associated Mollicutes through different species revealing a possible cospeciation pattern.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Brune A, Ohkuma M. Role of the termite gut microbiota in symbiotic digestion In Biology of termites: a modern synthesis. Dordrecht: Springer; 2010. 439-475 p.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

