Global Phosphoproteomic Analysis Reveals the Defense and Response Mechanisms of Jatropha Curcas Seedling under Chilling Stress
- PMID: 30626061
- PMCID: PMC6337099
- DOI: 10.3390/ijms20010208
Global Phosphoproteomic Analysis Reveals the Defense and Response Mechanisms of Jatropha Curcas Seedling under Chilling Stress
Abstract
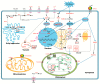
As a promising energy plant for biodiesel, Jatropha curcas is a tropical and subtropical shrub and its growth is affected by one of major abiotic stress, chilling. Therefore, we adopt the phosphoproteomic analysis, physiological measurement and ultrastructure observation to illustrate the responsive mechanism of J. curcas seedling under chilling (4 °C) stress. After chilling for 6 h, 308 significantly changed phosphoproteins were detected. Prolonged the chilling treatment for 24 h, obvious physiological injury can be observed and a total of 332 phosphoproteins were examined to be significantly changed. After recovery (28 °C) for 24 h, 291 phosphoproteins were varied at the phosphorylation level. GO analysis showed that significantly changed phosphoproteins were mainly responsible for cellular protein modification process, transport, cellular component organization and signal transduction at the chilling and recovery periods. On the basis of protein-protein interaction network analysis, phosphorylation of several protein kinases, such as SnRK2, MEKK1, EDR1, CDPK, EIN2, EIN4, PI4K and 14-3-3 were possibly responsible for cross-talk between ABA, Ca2+, ethylene and phosphoinositide mediated signaling pathways. We also highlighted the phosphorylation of HOS1, APX and PIP2 might be associated with response to chilling stress in J. curcas seedling. These results will be valuable for further study from the molecular breeding perspective.
Keywords: Jatropha curcas; chilling stress; phosphoproteomics; regulated mechanism; seedling.
Conflict of interest statement
The authors declare no conflict of interest.
Figures








Similar articles
-
Phosphoproteomic Analysis of Paper Mulberry Reveals Phosphorylation Functions in Chilling Tolerance.J Proteome Res. 2017 May 5;16(5):1944-1961. doi: 10.1021/acs.jproteome.6b01016. Epub 2017 Apr 13. J Proteome Res. 2017. PMID: 28357858
-
Osmoregulation is a crucial factor for methyl jasmonate to enhance chilling tolerance of Jatropha curcas L.Tree Physiol. 2025 Mar 28;45(4):tpaf037. doi: 10.1093/treephys/tpaf037. Tree Physiol. 2025. PMID: 40143413
-
Phosphoproteomics of cold stress-responsive mechanisms in Rhododendron chrysanthum.Mol Biol Rep. 2022 Jan;49(1):303-312. doi: 10.1007/s11033-021-06874-0. Epub 2021 Nov 6. Mol Biol Rep. 2022. PMID: 34743272
-
Phosphoproteomics: Protein Phosphorylation in Regulation of Seed Germination and Plant Growth.Curr Protein Pept Sci. 2018 Feb 13;19(4):401-412. doi: 10.2174/1389203718666170209151048. Curr Protein Pept Sci. 2018. PMID: 28190389 Review.
-
Revealing plant defense signaling: getting more sophisticated with phosphoproteomics.Plant Signal Behav. 2011 Oct;6(10):1469-74. doi: 10.4161/psb.6.10.17345. Epub 2011 Oct 1. Plant Signal Behav. 2011. PMID: 21897123 Free PMC article. Review.
Cited by
-
Chilling stress response in tobacco seedlings: insights from transcriptome, proteome, and phosphoproteome analyses.Front Plant Sci. 2024 May 28;15:1390993. doi: 10.3389/fpls.2024.1390993. eCollection 2024. Front Plant Sci. 2024. PMID: 38872895 Free PMC article.
-
Integration of transcriptomic and proteomic analyses of Rhododendron chrysanthum Pall. in response to cold stress in the Changbai Mountains.Mol Biol Rep. 2023 Apr;50(4):3607-3616. doi: 10.1007/s11033-022-08114-5. Epub 2022 Nov 23. Mol Biol Rep. 2023. PMID: 36418773
-
Integrative transcriptomic, proteomic, and phosphoproteomic analysis on the defense response to Magnaporthe oryzae reveals different expression patterns at the molecular level of durably resistant rice cultivar Mowanggu.Front Plant Sci. 2023 Jul 13;14:1212510. doi: 10.3389/fpls.2023.1212510. eCollection 2023. Front Plant Sci. 2023. PMID: 37521912 Free PMC article.
-
Autophagy deficiency confers freezing tolerance in Arabidopsis thaliana.BMC Plant Biol. 2025 Jul 30;25(1):994. doi: 10.1186/s12870-025-07066-9. BMC Plant Biol. 2025. PMID: 40739613 Free PMC article.
-
Deciphering the Proteome and Phosphoproteome of Peanut (Arachis hypogaea L.) Pegs Penetrating into the Soil.Int J Mol Sci. 2025 Jan 14;26(2):634. doi: 10.3390/ijms26020634. Int J Mol Sci. 2025. PMID: 39859350 Free PMC article.
References
-
- Thakur P., Kumar S., Malik J.A., Berger J.D., Nayyar H. Cold stress effects on reproductive development in grain crops: An overview. Environ. Exp. Bot. 2010;67:429–443. doi: 10.1016/j.envexpbot.2009.09.004. - DOI
-
- Levitt J. Chilling, Freezing and High Temperature Stresses. Responses of Plants to Environmental Stress. 2nd ed. Volume 1. Academic Press; London, UK: New York, NY, USA: 1980. p. 497.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

